High Levels of Antibodies to Plasmodium falciparum Liver Stage Antigen-1 in Naturally Infected Individuals in Myanmar
Article information
Abstract
Plasmodium falciparum liver stage antigen-1 (PfLSA-1) is one of the few antigens expressed exclusively in liver stage parasites. In this study, we evaluated the antibody responses against recombinant PfLSA-1 in naturally infected individuals in Myanmar. High levels of antibody responses (70.7%) were detected in 82 serum samples from 116 infected individuals, and IgG responses to PfLSA-1 principally composed of responses of IgG1 and IgG3 subclasses. These results show that PfLSA-1 elicits effective antibody responses in individuals infected with P. falciparum, and thus it could be not only an attractive candidate protein for vaccine development, but also a useful antigen for serodiagnosis of the infection.
The sporozoites of malaria parasites transmitted from the saliva of infected mosquitoes rapidly travel to the liver and invade hepatocytes, where they develop into the exoerythrocytic stage referred to as the tissue schizont. During this stage, the parasites express stage-specific antigens. Liver stage antigen-1 of Plasmodium falciparum (PfLSA-1) is a surface protein which is solely expressed in infected hepatocytes and is believed to play a role in liver schizogony and the release of merozoites [1,2]. PfLSA-1 induced specific humoral, cellular, and cytokine immune responses in infected individuals [3,4] and have been considered a vaccine candidate for P. falciparum, due to their antigenic and protection-including immunogenic properties [5,6]. In the current study, we evaluated antibody responses against PfLSA-1 in naturally infected individuals in Myanmar in order to expand our knowledge regarding the antigenic properties of PfLSA-1, and to evaluate its potential as an antigen for vaccine development and serodiagnosis of P. falciparum infection.
Blood samples were collected from inhabitants who visited the Wet-Won Station Hospital, in Pyin Oo Lwin Township, Mandalay Division, Myanmar, with clinical symptoms associated with malaria. Thin and thick blood smears were prepared from blood collected from individuals' fingertips for microscopic diagnosis. A local health unit employee read the Giemsa-stained blood smears, and treatment was administered to those individuals testing positive for malaria, on the basis of the guidelines established by The Department of Health of The Union of Myanmar. Additional blood samples of approximately 1 ml were collected from a total of 116 individuals in whom P. falciparum infection has been confirmed via microscopic examination, and these were utilized in this study. For negative controls, 50 serum samples from healthy volunteers were employed. All healthy volunteers worked at National Institute of Health, Korea Centers for Disease Control and Prevention, and had never been exposed to malaria nor visited malaria-endemic areas. Informed consent was acquired from all the participating individuals. The study protocol was approved by the Department of Health (Upper Myanmar) of The Union of Myanmar, and were carefully reviewed and approved by the Ethical Committees of the National Institute of Health, Korea Centers for Disease Control and Prevention.
The genomic DNA of P. falciparum was extracted from the patients' whole blood using a NucleoSpin Blood Kit (Macherey-Nagel GmbH & Co, Düren, Germany). Amplification of the C-terminal portion of PfLSA-1 which harbors major B- and T-helper epitopes [7] was conducted via polymerase chain reaction (PCR) using the forward primer 5'-GGATCCAGAAAAAAGGAACATGGAGATATATTAGCA-3' which harbored the Bam HI site at the 5' end and the reverse primer 5'-AAGCTTCTCATCCACGATCTGTAAAATTTCATTGTC-3' containing the Hind III site at the 5' end. PCR was conducted with AccuPower™ PCR Premix (Bioneer Co., Daejeon, Korea), 50 ng of purified genomic DNA, and 40 pmoles of reverse and forward primer sets. The reaction conditions were as follows: 94℃ for 10 min, 35 cycles of 1 min at 94℃, 1 min at 58℃, 1 min at 72℃ and a final extension step at 72℃ for 10 min. The PCR product was confirmed and purified with NucleoSpin Extract II kit (Macherey-Nagel GmbH & Co). The purified PCR product was ligated into pCR2.1-TOPO™ cloning vector (Invitrogen, Carlsbad, California, USA) and transformed into E. coli TOP10 (Invitrogen). Sequencing analysis was conducted with a BigDye Terminator Cycle Sequencing Ready Reaction Kit in an ABI 377 automatic DNA sequencer (Applied Biosystems, Foster City, California, USA) and the nucleotide and deduced amino acid sequences were analyzed with the SeqEd.V1.0.3 program of the DNASTAR package (DNASTAR, Madison, Wisconsin, USA).
To express recombinant PfLSA-1 (rPfLSA-1), the cloned PfLSA-1 was digested with Bam HI and Hind III and purified with a NucleoSpin Extract II kit (Macherey-Nagel GmbH & Co), cloned into pQE-30 expression vector (Qiagen, Valencia, California, USA) pretreated with the same restriction enzymes, and transformed into E. coli SG13009 cells (Qiagen). The selected clones were grown and induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). The bacteria were then suspended in native lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), sonicated on ice and centrifuged for 20 min at 4℃ at 12,000 g. The supernatant was collected and rPfLSA-1 was purified via nickel-nitrilotriacetic acid (Ni-NTA) chromatography (Qiagen). The purification and purity of rPfLSA-1 were analyzed via SDS-PAGE.
For western blot analysis, the purified rPfLSA-1 was separated on 10% SDS-PAGE gel, and the protein was transferred onto a nitrocellulose membrane. After the transfer, the membrane was cut into strips and blocked for 2 hr in 3% skim milk/phosphate buffered saline containing 0.02% Tween 20 (PBST) at room temperature. The strips were then washed 3 times with PBST and incubated for 4 hr in 1 : 100 diluted serum samples obtained from either normal or P. falciparum-infected individuals in PBST at room temperature. The strips were washed 3 times with PBST, added to 1 : 1,000 diluted peroxidase-conjugated anti-human IgG (Sigma, St. Louis, Missouri, USA) in PBST, and incubated for 3 hr at room temperature. For color development, 0.2% diaminobenzidine and 0.002% H2O2/PBS were used. IgG1, IgG2, IgG3, and IgG4 subclass antibodies were measured via enzyme-linked immunosorbent assay (ELISA). ELISA was conducted using subclass-specific monoclonal anti-human antibodies (Sigma; IgG1, clone 8c/6-39; IgG2, clone HP-6002; IgG3, clone HP-6050; and IgG4, clone HP-6025, respectively) as previously described [8].
The expression of the cloned C-terminal portion of PfLSA-1 was induced via the addition of IPTG and analyzed by SDS-PAGE followed by Coomassie blue staining. The molecular weight of the rPfLSA-1 was approximately 50 kDa, which is very consistent with the expected size of the protein on the basis of the gene sequence (Fig. 1). In order to analyze the antibody responses to rPfLSA-1 in naturally infected individuals, western blot analysis was conducted. A total of 82 serum samples (70.7%) among 116 sera from P. falciparum-infected patients were allowed to react with rPfLSA-1. Meanwhile, all the sera obtained from normal individuals (n = 50) did not react with the rPfLSA-1 (Fig. 2). These results collectively indicated that PfLSA-1 is highly immunogenic, and high levels of antibody responses are induced against rPfLSA-1 in individuals naturally infected with P. falciparum. In order to further determine the IgG subclasses which recognize rPfLSA-1, ELISA was also conducted. The patient sera reacted predominantly to IgG1 and IgG3 subclasses (Fig. 3).
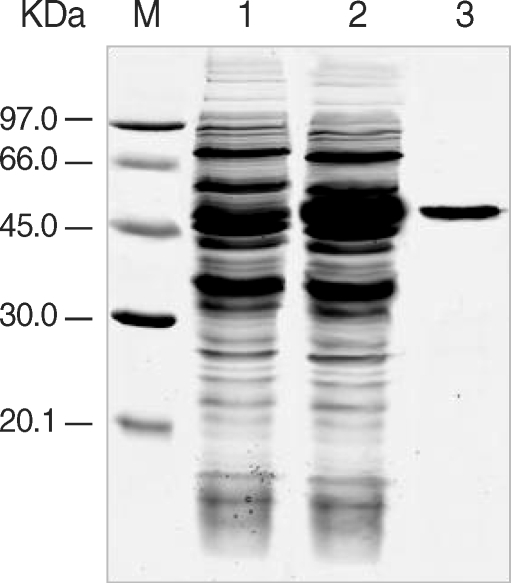
Expression and purification of rPfLSA-1. SDS-PAGE analysis of the expressed rPfLSA-1 protein. Lane M, molecular weight marker proteins; Lane 1, non-induced E. coli cell lysate; Lane 2, induced E. coli cell lysate; Lane 3, purified protein from induced E. coli cell lysate using Ni-NTA column.
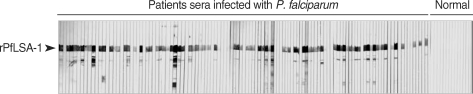
Western blot analysis of the purified rPfLSA-1 against the sera from patients naturally infected with P. falciparum in Myanmar and from normal people. Only representative reactions are shown.
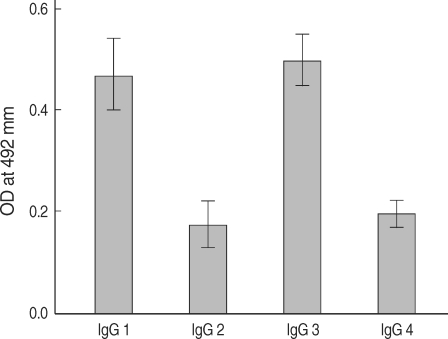
IgG subclass responses against rPfLSA-1 in the sera from patients naturally infected with P. falciparum in Myanmar.
High levels of antibody responses against a variety of exoerythrocytic stage proteins, including circumsporozoite protein (CSP), liver stage antigens (LSA-1 and LSA-3), and thrombospondin-related adhesive protein (TRAP) of P. falciparum in infected individuals have been identified and characterized [9-11]. Careful analysis of the antibody responses in malaria-infected individuals is crucial not only for our understanding of immune responses against the infection but also for the design of a vaccine or serodiagnostic method for malaria. Myanmar is a country with a high level of malaria endemicity. Although morbidity and mortality rates due to malaria have been gradually declining, owing to increased efforts to prevent and control the disease, Myanmar still contributes to approximately 60% of malaria deaths in Southeast Asia [12]. In the present study, high levels (70.7%) of antibody responses against PfLSA-1 were evoked in naturally infected individuals in Myanmar. This finding was consistent with the results of previous studies showing that a significant level of antibodies against PfLSA-1 can be detected in exposed or infected individuals with P. falciparum [10,13]. We also determined that IgG responses to PfLSA-1 were principally composed with responses of IgG1 and IgG3 subclasses. Our findings for IgG subclass antibodies to PfLSA-1 are similar to those previously reported [13,14]. In general, subclasses IgG1 and IgG3 are known to mediate opsonization and the complement fixation of microorganisms, and have been associated with antibody-mediated protective immunity against blood stage malaria [15]. Previous studies have also shown that IgG1 and IgG3 antibodies predominate in protected adults, whereas IgG2 and IgG4 predominate in non-protected children and adults who have suffered from P. falciparum primary attack [16-18]. Indeed, individuals exposed to either natural or experimental malarial infections generate immune responses (proliferative T-cells, cytokines, and antibodies) to LSA-1 protein or peptides that have been associated with complete protection or reduced parasitemia upon subsequent exposure [3,19,20]. Our findings of dominant antibody responses of IgG1 and IgG3 against PfLSA-1 in naturally infected individuals indicate that this may potentially contribute to the induction of protective effects via cell-mediated mechanisms. However, further studies involving the significance of immune responses and the vaccine potential of PfLSA-1 are needed.
ACKNOWLEDGEMENTS
This work was supported by a grant from the Korea Health 21 R & D Project, Ministry of Health and Welfare, Republic of Korea (03-PJ1-PG1-CH01-0001).