Morphologic and Genetic Identification of Taenia Tapeworms in Tanzania and DNA Genotyping of Taenia solium
Article information
Abstract
Species identification of Taenia tapeworms was performed using morphologic observations and multiplex PCR and DNA sequencing of the mitochondrial cox1 gene. In 2008 and 2009, a total of 1,057 fecal samples were collected from residents of Kongwa district of Dodoma region, Tanzania, and examined microscopically for helminth eggs and proglottids. Of these, 4 Taenia egg positive cases were identified, and the eggs were subjected to DNA analysis. Several proglottids of Taenia solium were recovered from 1 of the 4 cases. This established that the species were T. solium (n=1) and T. saginata (n=3). One further T. solium specimen was found among 128 fecal samples collected from Mbulu district in Arusha, and this had an intact strobila with the scolex. Phylegenetic analysis of the mtDNA cox1 gene sequences of these 5 isolates showed that T. saginata was basal to the T. solium clade. The mitochondrial cox1 gene sequences of 3 of these Tanzanian isolates showed 99% similarity to T. saginata, and the other 2 isolates showed 100% similarity to T. solium. The present study has shown that Taenia tapeworms are endemic in Kongwa district of Tanzania, as well as in a previously identified Mbulu district. Both T. solium isolates were found to have an "African/Latin American" genotype (cox1).
INTRODUCTION
Between 2005 and 2009, a Korean NGO, named Good Neighbors International, funded a survey of neglected tropical diseases in Tanzania, in particular, helminth infections. The aim of this collaborative project was to promote health through control of parasitic infections among school children. Various parasitic helminth infections have been reported in Tanzanians, i.e., ascariasis, trichuriasis, hookworm infections, strongyloidiasis, enterobiasis, trichinosis, taeniasis, hydatidosis, echinococcosis, and sparganosis. Previous studies in this population have reported cestode infections, including taeniasis, hydatidosis, echinococcosis, and sparganosis. The first reported case of hydatid disease involved a Massai patient, who was found to have multiple unilocular cysts [1]. The prevalence of this disease increased from 2.4% in 1967 to 5.6% in 1971, and ultrasound scanning of 959 individuals revealed an average prevalence of 1.4% [2]. Schmid and Watschinger reported cases of sparganosis in the Massai tribes, and performed over 20 operations on Massai patients [3]. Dodoma, Kilimanjaro, Mwanza, and the Ukara islands in Lake Victoria showed the highest prevalence of human taeniasis nationwide, with most cases involving Taenia saginata. To date, however, there have been no clear reports of human cysticercosis/taeniasis infection, in particular, adult Taenia solium in Tanzania.
Taeniasis and cysticercosis remain a significant public health concern in parts of Asia, Africa, Eastern Europe, and Central and South America. T. solium cysticercosis has also been reported in pigs and humans in many countries in western and central Africa. The status of human taeniasis in Tanzania is exceptional in terms of scientific reporting. In Nigeria, the prevalence of porcine cysticercosis and human taeniasis is as high as 20.5% and 8.6%, respectively. In Togo and Benin, the prevalence of human cysticercosis is 2.4% and 1.3%, respectively. In central Africa, porcine and human cysticercosis are hyperendemic in Rwanda, Burundi, Congo, and Cameroon [4]. The prevalence of cysticercosis among African pigs has been estimated to be as high as 5.7-17.4% [5], according to an investigation of muscle biopsy specimens obtained from abattoirs at various localities. However, the true prevalence is probably higher, since these data were based on the results of visual inspection with the naked eye or lingual palpation. In 2003, the Cysticercosis Working Group in Eastern and Southern Africa published a 'Declaration on Taenia solium cysticercosis/taeniosis' [6].
In 2008 and 2009, the present authors conducted field investigations for helminth infections and T. solium in 2 districts of mid-Tanzania: (i) Kongwa, in the Dodoma region; and (ii) Mbulu, in the northwest Arusha region. The present study was conducted within the context of these investigations, and focused on identification of helminth and cestode cases. This was achieved by microscopic examination for the presence of eggs in the stool. Taenia-specific dot-ELISA coproantigen tests, morphologic observation of adult worms, and multiplex PCR and nucleotide analyses for sequence variations were used for the identification of human Taenia tapeworms.
MATERIALS AND METHODS
Specimens and stool examination
In 2008 and 2009, a total of 1,057 stool samples were collected from Kongwa district of Tanzania; Ijaka (n=199); Pembamoto (n=730), and Khaday (n=128) (Table 1). Of these, 929 were examined using the Kato-Katz method, and the remaining 128 samples were examined using the scotch-tape anal swab. Patients with Taenia tapeworms discharged adult worms after treatment with niclosamide or praziquantel with purgation. The morphologic features of the proglottids were compatible with those of human Taenia tapeworms. Mature proglottids were examined with acetocarmine staining. Scolices were pressed between 2 slide glasses and examined using light microscopy.

Helminth egg positive rates for inhabitants and students in Kongwa and Mbulu districts of Tanzania using the Kato-Katz method
Five Taenia cases from Mbulu and Kongwa districts were identified. The code number 1614 (a 15-year-old male) living in Mbulu discharged an entire worm after treatment with praziquantel and purgation. The code number 1658 (a 13-year-old female) from Ijaka discharged 9 proglottids following the same treatment. The code numbers 1673 (a 15-year-old male), 1674 (a 40-year-old male), and 1677 (a 14-year-old male), all from Pembamoto, were found to be egg positive using either the scotch-tape anal swab or the Kato-Katz technique. The Taenia eggs were isolated from the stools and examined by multiplex PCR.
Dot ELISA
Dot ELISA was performed as described previously [7]. Dot ELISA were dotted with a 2-µl (0.15 µg/ml) concentration of anti-T. solium IgG in a PBS coating on the nitrocellulose. The nitrocellulose was dried at room temperature for 1 hr, and then blocked with 5% dry skimmed milk in PBS for 1 hr. The fecal sample was prepared by adding an equal weight volume ratio of PBS containing 0.3% Tween 20. This was shaken vigorously to form a slurry, and then centrifuged at 3,500 rpm for 30 min at room temperature. Fecal supernatants were prepared, and the nitrocellulose was incubated in these supernatants for 30 min at room temperature. The nitrocellulose was then washed with water to remove any fecal material and incubated immediately in a 1:1,000 dilution of peroxidase-conjugated anti-T. solium IgG for 30 min. The nitrocellulose was then washed 3 times in water and incubated in a substrate (10 mg tablet DAB) in 25 ml of PBS with 10 µl of 30% H2O2 for precisely 2 min. The nitrocellulose was then washed with water and dried for 30 min (for the presence of any visible dot).
Multiplex PCR and DNA sequence analyses of the Taenia eggs
Multiplex PCR was performed as described previously [8]. The purified PCR-amplified fragments of the cox1 were then directly sequenced. The mitochondrial cox1 gene was targeted in PCR amplification. The PCR primers were T1F (5'-ATA TTT ACT TTA GAT CAT AAG CGG-3') and TR1 (5'-ACG AGA AAA TAT ATT AGT CAT AAA-3') for cox1, which yield a 349-bp product. The second PCR reaction and sequencing primers were T1F1 (5'-TTT AGT TTA TTA ATT CGT GTT AAT-3') and T1R1 (5'-ATA AAA TTA ATA GAA CTA AAA ATA-3'). The primer walking method was employed to obtain direct sequences for each of the amplified fragments. Cyclic sequencing from both ends of the fragments was performed using a Big-Dye Terminator sequencing kit (Applied Biosystems Co., Grand Island, New York, USA), and the reaction products were electrophoresed on an automated DNA sequencer (model 3730KL, Applied Biosystems). The sequences were assembled and aligned using the Bioedit program (version 5.0.6, BIOSOFT, Ferguson, Missouri, USA). Using BLAST searches, the sequencing regions were identified by comparing them with those of Taenia tapeworms deposited in the GenBank database. Molecular identification of the Taenia tapeworm specimens was based upon a comparison with the nucleotide sequences of the cox1 genes of T. solium (GenBank No. AB086256), T. saginata (Genbank No. AY684274), and T. asiatica (GenBank No. AF445798). Phylogenetic analyses were performed using the neighbor-joining (NJ), maximum-parsimony (MP), and minimum-evolution (ME) methods, and the Mega 4.0 program [9]. NJ analysis was performed using a distance matrix calculated using the Kimura 2 parameter method. Bootstrap analysis was performed with 3,000 replications.
RESULTS
Helminth infections in Kongwa and Mbulu
The overall prevalence of parasitic helminths was 6.3% (67/1,057) (Table 1). The overall prevalence of Taenia eggs was 0.5% (5/1,057), 4 cases in Kongwa and 1 case in Mbulu. In Pembamoto and Ijaka, 5 other types of helminth eggs were found; Trichuris trichiura, Ascaris lumbricoides, Enterobius vermicularis, Hymenolepis diminuta, and Hymenolepis nana. In Mbulu, only 1 case, 0.8% (1/128), was found to have a helminth infection which involved Taenia infection. He discharged an entire tapeworm strobila after treatment with praziquantel and purgation with hydrated magnesium sulfate and a large volume of water. From Ijaka, a 14-year-old girl passed 9 proglottids in her stool after treatment with niclosamide and purgation. In the remaining 3 cases, only eggs were collected from the feces.
Dot ELISA
A total of 364 fresh fecal samples from Pembamoto and Ijaka were examined using dot ELISA. A total of 8 cases were found to be T. solium positive. Of these, 4 cases were proven by the presence of eggs in feces or recovery of proglottids after treatment. In the 4 remaining cases, no parasite eggs were detected.
Differential diagnosis of Taenia tapeworms by multiplex PCR
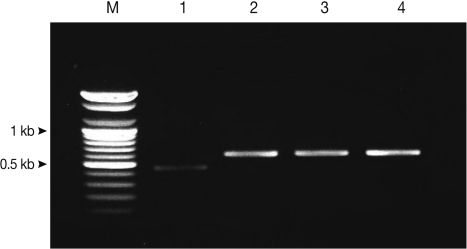
Using multiplex PCR, a 629-bp diagnostic band was detected in the T. saginata samples of code numbers 1673, 1674, and 1677. In the T. solium samples, a 474-bp diagnostic band was detected in case number 1629. In the egg positive cases, multiplex PCR for copro-DNA clearly demonstrated the 629-bp and 474-bp diagnostic bands of T. solium and T. saginata (Fig. 1).
Observation of adult Taenia solium
Morphology
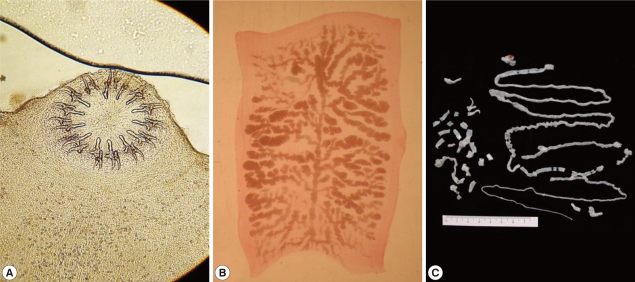
The whitish-yellow adult tapeworm discharged by code number 1614 was 1.5 m in length and consisted of 199 proglottids with unilateral genital pores. The morphological features of these proglottids were compatible with T. solium. The scolex was rectangular in shape and 1.2 mm in width with 2 rows of 30 hooklets as well as 4 suckers. Gravid proglottids were examined using light microscopy with carmine staining of the press-and-fixed specimen (acetic acid-formalin-alcohol solution). The proglottids from code numbers 1614 and 1658 had 10 or fewer uterine main branches unilaterally, and were thus identified as T. solium. The eggs were ball-shaped and their average size was 33.5±1.0 µm (n=30) (Fig. 2).
DNA genotype
The mitochondrial cox1 sequences of 3 Tanzanian isolates (1673, 1674, and 1677) showed a 99% similarity to reference sequences of the Belgium origin T. saginata (GenBank No. AY195858). Two isolates (1614 and 1658) showed a 100% similarity to the reference sequences of Tanzanian origin T. solium (GenBank No. AB066493), based on Kimura's 2 parameter model. Phylogenetic analysis of the mtDNA cox1 sequences for the 5 isolates showed that T. saginata was basal to the T. solium clade. Tree topology was identical with very high confidence values (bootstrap values of 100%, 99%, and 100% in NJ, MP, and ME, respectively) for 2 major branches, representing T. sagianta and T. solium, respectively (Fig. 3). Analysis of the cox1 sequences revealed that 3 of the Tanzanian specimens were T. saginata and 2 were T. solium. Both of these T. solium isolates were classified as having an "African/Latin American" and a "non-Asian" genotype.
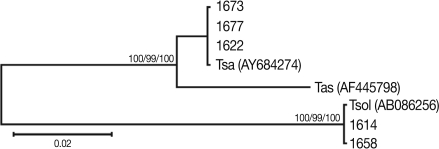
Phylogenetic trees of human Taenia tapeworms based on partial nucleotide sequences of cox1 gene fragment inferred from neighbor-joining (NJ), maximum parsimony (MP), and minimum evolution (ME) analyses. The number at each node is the bootstrap confidence value obtained after 3,000 iterations. The scale bar represents the estimated number of nucleotide substitutions per nucleotide site. Tsa, T. saginata; Tas, T. asiatica; Tsol, T. solium. Code no. 1618, 1629, 1658, 1673, and 1677 Tanzanian isolates of Taenia species.
DISCUSSION
Research has shown that the major parasitic infections in Tanzania are malaria, trypanosomiasis, schistosomiasis, bancroftian filariasis, and onchocerciasis. However, very little is known about the distribution and prevalence of protozoan and other helminth parasites. In the present study, T. trichiuria, A. lumbricoides, E. vermicularis, H. nana, H. diminuta, T. saginata, and T. solium were observed in Kongwa (Pembamoto and Ijaka) and Mbulu (Khaday) districts of Tanzania. In Kongwa and Mbulu districts, T. solium was found to be endemic. T. solium metacestodes have been reported as an emerging and increasing problem amongst small-holder pig populations in northern and southern highlands of Tanzania [10]. Porcine cysticercosis was identified in pigs in the northern highland district of Mbulu, having first been detected in pigs exported to Kenya [10,11]. The prevalence of porcine T. solium cysticerci in Mbulu district has been estimated to be as high as 10.6% [12] or 17.4% [13], with a prevalence of 5.1-16.9% in the southern highlands [5].
Interviews with the inhabitants of the northern highlands of Tanzania [14] have indicated that human cysticercosis may be highly prevalent. However, few surveys of human infections have been undertaken in Tanzania, even though a high prevalence of porcine cysticercosis has been found at various localities across the country. The high prevalence of porcine cysticercosis indicates the presence of a large number of reservoir human Taenia tapeworm carriers, who are destined to infect pigs as well as humans. A serological survey (i.e., ELISA) would be appropriate in endemic areas, such as Mbulu and some other areas [8,12]. According to local public health tapeworm experts, T. solium may be common among pork eaters, such as the Chaga (Kilimanjaro) and the Wanyiramba (Singida), while T. saginata may be common amongst beef eaters, such as the Massai (Arusha) and the Wagogo (Dodoma). Detailed parasitological evaluation is considered to be necessary among these people and regions.
In the present study, T. solium (n=2) and T. saginata (n=3) were identified in samples collected from Kongwa and Mbulu districts of Tanzania. Their morphologic characteristics were documented, and the egg DNA was subjected to genotyping of the cox1 sequence and characterization using multiplex PCR.
ACKNOWLEDGMENTS
The authors thank Dr. Ursuline Nyandindi (Ministry of Education) as well as Dr. Ali Mzige and Dr. Rafael Kalinga (Ministry of Health, Dar Es Salaam, United Republic of Tanzania) for their help with the "Korea-Tanzania Collaborative Project on Health Promotion through Parasite Control among School Children, 2005-2009" which was supported by Good Neighbors International, Korea. Technological support for this work was provided in 2011 through a research grant from Chungbuk National University, Korea. The parasite materials used in this study were provided by the Parasite Resource Bank of Korea, National Research Center (R21-2005-000-10007-0), Republic of Korea.