CD8+ T-cell Activation in Mice Injected with a Plasmid DNA Vaccine Encoding AMA-1 of the Reemerging Korean Plasmodium vivax
Article information
Abstract
Relatively little has been studied on the AMA-1 vaccine against Plasmodium vivax and on the plasmid DNA vaccine encoding P. vivax AMA-1 (PvAMA-1). In the present study, a plasmid DNA vaccine encoding AMA-1 of the reemerging Korean P. vivax has been constructed and a preliminary study was done on its cellular immunogenicity to recipient BALB/c mice. The PvAMA-1 gene was cloned and expressed in the plasmid vector UBpcAMA-1, and a protein band of approximately 56.8 kDa was obtained from the transfected COS7 cells. BALB/c mice were immunized intramuscularly or using a gene gun 4 times with the vaccine, and the proportions of splenic T-cell subsets were examined by fluorocytometry at week 2 after the last injection. The spleen cells from intramuscularly injected mice revealed no significant changes in the proportions of CD8+ T-cells and CD4+ T-cells. However, in mice immunized using a gene gun, significantly higher (P<0.05) proportions of CD8+ cells were observed compared to UB vector-injected control mice. The results indicated that cellular immunogenicity of the plasmid DNA vaccine encoding AMA-1 of the reemerging Korean P. vivax was weak when it was injected intramuscularly; however, a promising effect was observed using the gene gun injection technique.
Intramuscular or gene gun-based injections of plasmid DNA expression vectors, i.e., DNA vaccines, induce corresponding protein expression in vivo and generate humoral and cellular immune responses against various infectious agents [1]. DNA vaccines are simple, inexpensive, and heat-stable, and can induce protective immunity without using live organisms, replicating vectors, or harmful adjuvants [2]. Significant progress has recently been made in the development of malaria DNA vaccines targeting the pre-erythrocytic and erythrocytic stage antigens. Circumsporozoite proteins (CSP), merozoite surface proteins (MSP), Duffy-binding protein (DBP), and apical membrane antigens (AMA) have been used as antigens [2-4].
Apical membrane antigen 1 (AMA-1), previously known as Pf83 and Pk66 in Plasmodium falciparum, is a micronemal protein of apicomplexan parasites and is essential during the invasion of malarial parasites into host cells [5]. There is a single gene for this antigen in all Plasmodium species [5]. AMA-1 is a good vaccine candidate because it can have profound parasite-inhibitory effects in vitro and in animal models [5]. Several studies have examined the vaccine potential of AMA-1 against P. falciparum and other human-infecting malaria [6,7]. However, relatively little has been studied on AMA-1 vaccines against Plasmodium vivax [8,9].
Since important issues were raised about P. vivax, including its global burden, drug resistance, severity of the disease, prevalence of relapse and recrudescence, and problems of coinfection with P. falciparum, there is a renewed interest in the development of a P. vivax vaccine [10]. A modest number of P. vivax vaccine candidates, including CSP, MSP, and DBP, have been tested in pre-clinical trials in rodents [11], and several CSPs and an ookinete surface antigen (Pvs25) were assessed in phase I clinical trials [10]. Vaccination with P. vivax AMA-1 (PvAMA-1) was attempted in primates [8], and PvAMA-1 was shown to elicit differentiation of dendritic cells in naturally infected vivax malaria patients [9]. However, few studies have been reported on plasmid DNA vaccines encoding PvAMA-1 [3]. The present study aimed to construct a DNA plasmid vaccine encoding AMA-1 of the reemerging P. vivax in the Republic of Korea (=Korea), and to preliminarily observe its cellular immunogenicity in recipient BALB/c mice.
Six-week-old female BALB/c mice (Koatech, Pyeongtaek, Gyeonggi-do, Korea) were supplied with food and water sterilized by irradiation and autoclaving. All animal procedures were performed according to the approved protocols and institutional recommendations for the proper use and care of laboratory animals, Seoul National University College of Medicine.
Blood samples were collected from 17 P. vivax patients (designated SKPV1 through SKPV17) diagnosed at the Department of Parasitology and Tropical Medicine, Seoul National University College of Medicine, Seoul, and Paju Medical Center, Paju, Gyeonggi-do (Province), Korea between 1995 and 2000, and frozen at -80℃. The level of parasitemia ranged from 500 to 6,200 parasites per µl blood. The genomic DNA of P. vivax was extracted using the QIAmp DNA Mini kit (Qiagen, Hilden, Germany). After ethanol precipitation, it was dissolved in distilled water and kept at -80℃. After PCR amplification of AMA-1, its nucleotide sequences were compared with those of the Salvador strain (Sal-1) (GenBank accession number; AF063138). The primer sets were based on the oligonucleotide sequences of PvAMA-1 [12]. One µl of genomic DNA and 20 pmol each of forward and reverse primers were added to the PCR premix (Bioneer, Seoul, Korea). The PCR products were amplified in 35 cycles using a GeneAmp PCR System 9600 DNA thermal cycler (PE Applied Biosystems, Forster City, California, USA). The PCR conditions were as follows: denaturation at 95℃ for 1 min, annealing at the indicated temperature for 1 min, and extension at 72℃ for 1 min. The DNA band visualized by ethidium bromide staining after electrophoresis was extracted using the QIAEX II gel extraction kit (Qiagen).
The PCR products of AMA-1 were subcloned using the pCR2.1 cloning vector of a TOPO Cloning Kit (Invitrogen, Carlsbad City, California, USA). Escherichia coli, strain JM109, was used as the host for transformation. When target fragments from positive clones were confirmed, the plasmid was prepared using the miniprep kit (Qiagen) and sequenced using a model ABI Prime 377 Automatic Sequencer (PE Applied Bio-systems). The gene UBpcDNA encoding the mutant ubiquitin, whose C-terminal Gly residue was replaced by Ala (G74A), originated in the liver of BALB/c mice and was inserted into the Nhe I and Xho I sites of pcDNA 3.1(-) vector, which was supplied by the Department of Parasitology, Graduate School of Medical Sciences, Kyushu University, Japan [13].
For construction of UBpcAMA-1, the miniprep products of the DNA-TA vector for AMA-1 were digested with Xho I and Apa I enzymes, and ligated to the 3' of the gene encoding mutant ubiquitin cDNA in the frame and inserted into the Apa I site of pcDNA3.1(-) attached C-terminal His tag (Fig. 1A). AMA-1 protein expression in COS7 cells (Korea Cell Line Bank, Seoul, Korea) transfected with UBpcAMA-1 plasmid was confirmed by immunofluorescence stain (Fig. 1B) and western blot (Fig. 1C). COS7 cells were cultured in DMEM medium supplemented with 10% FBS (GIBCO BRL, Grand Island, New York, USA) in a 5% humidified CO2 incubator at 37℃. At 24 hr after the transfection, 10 µM MG132, an inhibitor of proteasomes, was used and, at 48 hr, cells were harvested and lysed by the addition of 200 µl lysis buffer.
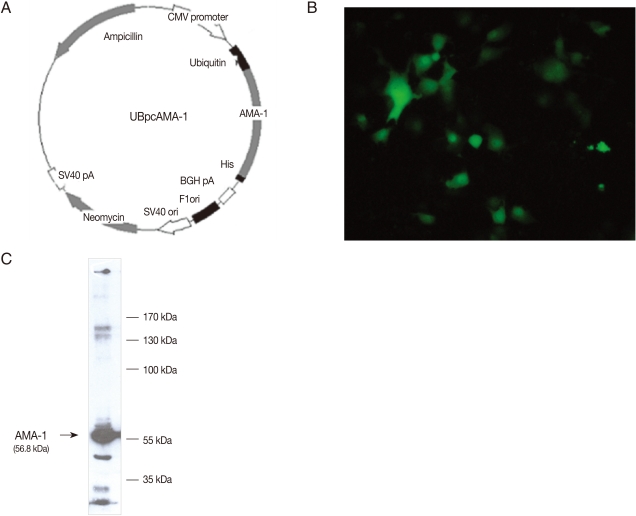
Expression of UBpcAMA-1 in mammalian COS7 cells. (A) A map showing the construction of the plasmid using the mammalian expression vector pcDNA 3.1(-), including the AMA-1 insert. The plasmid miniprep products from DNA-ubiquitin fused vector pcDNA 3.1(-) were cut by enzyme digestion with Xho I and Apa I. The antigen was cloned into the pcDNA 3.1(-) vector, and the expression plasmid was constructed. The vector was inserted with the mutant ubiquitin gene, so that the expression plasmid was expected to be generated by the ubiquitin-proteasome pathway in mammalian cells. (B) COS-7 cells transfected with UBpcAMA-1 using lipofectamine after immunofluorescence staining. The AMA-1 antigen was successfully expressed in cultured COS-7 cells. (C) Western blot analysis of the protein expression of the plasmid DNA in transfected COS7 cells. The PvAMA-1 recombinant protein was approximately 56.8 kDa.
DNA vaccine injection was performed either intramuscularly (10 µg vaccine in 100 µl saline) into the quadriceps muscle or with a gene gun into the epidermis [2]. Mice, 3-5 in each group, were anesthetized with sodium pentobarbital (75 mg/kg) and vaccinated a total of 4 times at 2 week intervals. For gene gun injection, the expression plasmid was precipitated onto 1.6 mm diameter gold particles (12.5 mg particles in 100 µl 0.05 M spermidine mixed with 100 µl of 100 µg plasmid), and the plasmid-gold particles were resuspended in 6 ml of ethanol and coated onto the inner surface of a tube. The plasmid DNA was delivered to the shaved abdominal skin of mice in 1 shot using the Helios Gene Gun (Bio-Rad, Hercules, California, USA) at a helium pressure of 300 p.s.i. For comparison, groups of mice were immunized with 2 µg UB vector (control group), 2 µg UBpcAMA-1 (AMA-1 group), 1 µg UBpcAMA-1 plus 1 µg UBpcIL-12 (AMA-1 + IL-12 group), or 2 µg UBpcIL-12 (IL-12 group) per shot.
Phenotype changes of splenocytes before and after vaccination were examined using flow cytometry at 2 weeks after the final boosting. Spleens were removed and gently crushed through a stainless steel mesh. After lysis of erythrocytes and washing for 10 min at 1,500 rpm, splenocytes were suspended in complete RPMI 1640 media. For fluorescence-activated cell sorter (FACS) analysis, cells were adjusted to 1×107 cells per ml, and 100 µl (1×106 cells) of this cell suspension was stained with specific antibodies. To avoid non-specific binding, purified rat anti-mouse CD16/32 (Fcγ III/II receptor) mAb (eBioscience, San Diego, California, USA) was added before labeling with specific antibodies. For phenotype determination, the following mAbs were used: fluorescein isothiocyanate-conjugated anti-mouse mAb against CD3+ T-cells (CD3e, clone 145-2C11) (eBioscience), Cy5-conjugated anti-mouse mAb against CD8+ T-cells (CD8a LY-2, clone 53-6.7) (eBioscience), and PE-conjugated anti-mouse mAb against CD4+ T-cells (CD4 L3T4, clone GK1.5) (eBioscience). Fluorescence was quantified using BD FACS Calibur Flow Cytometer (BD science, San Jose, New Jersey, USA).
Data were compared using the Mann-Whitney U test (SPSS Inc., Chicago, Illinois, USA) which is appropriate for testing small numbers of samples. A P-value of <0.05 was considered statistically significant.
Little polymorphism was observed in PvAMA-1 gene among the 17 reemerging Korean isolates (data not shown). Also, the nucleotide sequence of SKPV1, one of the 17 isolates, showed over 99% homology with other previously reported Korean isolates (SK-G, SK-A, SKOR-67, SKOR-68) and over 98% homology with foreign strains, Sal-1 (El Salvador) and PH-84 (the Philippines) (data not shown). The amino acid sequence homology was over 97% with Sal-1, over 98% with SK-G, SKOR-67, SKOR-68, and PH-84, and over 99% with SK-A.
The eukaryotic expression plasmid, UBpcAMA-1, was constructed as described in Fig. 1A. The mammalian expression vector, pcDNA 3.1(-), was inserted with AMA-1 and mutant ubiquitin genes, and thus the expression plasmid was generated by the ubiquitin-proteasome pathway in mammalian cells. The plasmid miniprep products from DNA-ubiquitin fused vector pcDNA 3.1(-) were cut by digestion with Xho I and Apa I enzymes. The DNA fragments of PCR products were subcloned into TA vectors and the genes encoding these antigens were sequenced. Then, the antigens were cloned into the pcDNA 3.1(-) vector and the expression plasmids were constructed (Fig. 1A). Immunofluorescence staining revealed brilliant cytoplasmic expression of AMA-1 in COS7 cells transfected with UBpcAMA-1 plasmid (Fig. 1B). We also confirmed the expression of expected proteins by western blot analysis. The immunoblots showed a large protein band of about 56.8 kDa present in COS7 cells transfected with UBpcAMA-1 (Fig. 1C).
The spleens of intramuscularly or gene gun immunized mice enlarged notably compared with those of the unimmunized controls. In particular, the spleen weight of the gene gun injected mice almost doubled that of the unimmunized controls (data not shown). FACS analysis of spleen cells harvested 2 weeks after the final immunization with AMA-1 or AMA-1 plus IL-12 DNA vaccines showed notable differences in the proportions of CD4+ and CD8+ T-lymphocytes between the intramuscular injection versus the gene gun (epidermis) injection (Fig. 2). In mice immunized intramuscularly (n=5 for each group) with AMA-1 alone or AMA-1 plus IL-12 DNA vaccines, the proportions of T-cell subsets (17.9±0.47% or 15.5±0.79% for CD8+ T-cells and 27.6±0.84% or 27.7±0.72% for CD4+ T-cells) did not show any significant changes (P>0.05), compared with control mice immunized only with UB vector (16.2±0.78% for CD8+ T-cells and 26.5±0.35% for CD4+ T-cells) (Fig. 2). By contrast, in mice injected with a gene gun (n=3 for each group), significantly higher proportions of CD8+ T-cells were found in those immunized with AMA-1, IL-12, or AMA-1 plus IL-12 DNA vaccines (24.9±0.44%, 20.8±0.61, or 24.2±1.56%, respectively), compared with control mice immunized only with UB vector (19.2±0.66%) (Fig. 2). However, even in mice injected with a gene gun, the proportions of CD4+ T-cells did not change significantly in AMA-1 alone or AMA-1 plus IL-12 injection groups (28.1±5.66% or 36.9±4.76%, respectively), compared with control mice immunized with UB vector (30.3±0.55%) (Fig. 2).

Changes of CD8+ and CD4+ T-lymphocyte population in PvAMA-1 DNA vaccine-immunized mice by intramuscular (IM) or gene gun injection methods. Immunization was performed a total of 4 times at 2 weeks interval with PvAMA-1 DNA vaccine alone or in combination with IL-12 DNA vaccine (n=5 mice for IM and n=3 for gene gun injection). Splenocytes were harvested 2 weeks after the final immunization and changes of CD4+ and CD8+ T-lymphocyte proportions were determined by flow cytometric analysis. Significant increases (P<0.05) of CD8+ T-cell populations were found in PvAMA-1 DNA vaccine alone and PvAMA-1 plus IL-12 DNA vaccine immunized groups compared with the controls injected with the gene gun.
In the reemerging vivax malaria in Korea, little polymorphism has been found in 18S rRNA [14] and AMA-1 [15,16] but a considerable degree of variation has been found on merozoite surface protein-3α [17]. Control programs have been in operation but no vaccine development has been reported. To our knowledge, this is the first report of a vaccine candidate targeting the reemerging vivax malaria in Korea.
The AMA-1 of P. vivax and P. falciparum has 3 extracellular domains (designated domains I, II, and III in N- to C-terminal order) [18]. The AMA-1 of P. vivax can have significant nucleotide sequence polymorphism at domain I, as seen in Myanmar isolates [19]. However, PvAMA-1 has little polymorphism at domain II, and has, therefore, been highlighted as a potential vaccine candidate [20]. The AMA-1 of the reemerging Korean P. vivax isolates has 2 genotypes and show little polymorphism [15,16]. The present study also revealed little genetic polymorphism in the PvAMA-1 of the reemerging Korean isolates. The sequence was over 98-99% homologous to the previously reported Korean (SK-A, SK-G, SKOR-67, and SKOR-68) or foreign isolates (Sal-1, PH-84, CH-05A, CH-10A, PNG, and INDO) [15,16]. Therefore, we considered that PvAMA-1 DNA vaccine is a potential vaccine candidate for the reemerging Korean P. vivax.
To confirm in vitro expression of AMA-1, we selected a eukaryotic expression system using COS7 mammalian cells for transfection with the plasmid vector UBpcAMA-1. Ubiquitin was used to fuse AMA-1 with the vector [13]. In this vector, mutant ubiquitin was expected to conduct antigen presentation to MHC class I molecules and to activate CD8+ T-cells [21]. In our study, ubiquitin-fused AMA-1 was successfully expressed after using a proteasome inhibitor, MG132, and confirmed as a 56.8 kDa protein by western blotting.
Despite the advantages of plasmid DNA vaccines, their applicability is generally limited by their poor immunogenicity and protective capacity [3]. One of the factors influencing the poor immunogenicity of DNA vaccines is a lowered efficacy of antigen expression [4]. Genes cloned from a pathogenic organism may not be efficiently translated in a heterologous host expression system as a consequence of codon bias displayed between different species [4]. One potential approach to enhance the immunogenicity of plasmid DNA vaccines is to maximize pathogenic protein expression by changing the codon usage of the gene of interest to reflect that of the gene of the transfected mammalian host cells [4,22,23]. In P. falciparum CSP, P. yoelii CSP, and P. yoelii hepatocyte erythrocyte protein (PyHEP17) DNA vaccine models, mammalian codon optimization enhanced expression of target proteins in vitro and antibody responses in immunized mice. However, codon optimization did not enhance T-cell responses or protective immunity [4]. The effect of codon optimization of DNA vaccines may vary depending on the antigen, pathogen, or host system, and should be tested on a case-by-case basis. In our study, we did not use codon optimization.
An important result of our preliminary study was that immunogenicity of PvAMA-1 DNA vaccine differed between intramuscular and gene gun immunization methods and that the gene gun immunization resulted in enhanced CD8+ T-cell responses of recipient mice. This result was in accordance with previous reports on a plasmid DNA vaccine encoding a cytotoxic T-lymphocyte epitope or ovalbumin (OVA) [24] and a DNA vaccine expressing a hemagglutinin antigen from an H5N1 influenza virus [25]. The gene gun method resulted in more reliable and reproducible results than the intramuscular injection method for DNA vaccination in inducing effective and consistent immune responses in animal models [24,25]. The difference in reproducibility may be related to the difference in antigen presentation mechanisms between the 2 methods [23]. In the gene gun system, the plasmid DNA is injected into the epidermis where the most important antigen presenting cells are epidermal dendritic cells (Langerhans cells). In the muscle, myocytes do not work as primary antigen presenting cells [22].
We regret that the actual cell numbers of different T-cell phenotypes in the mouse spleen were not obtained in fluorocytometric analyses. However, we observed that the spleens of immunized mice were enlarged, without exception, compared to the unimmunized controls. Therefore, it is for sure that the significant increases of CD8+ T-cell proportions represent substantial increases in the actual numbers of CD8+ T-cells in the immunized mice.
Conclusively, in our study, a plasmid DNA vaccine encoding AMA-1 of the reemerging Korean P. vivax has been successfully constructed and significantly higher (P<0.05) proportions of splenic CD8+ cells were observed in mice subcutaneously immunized using a gene gun compared to UB vector-injected non-immunized controls. Further studies on the efficacy of the gene gun technique will be useful for the development of P. vivax DNA vaccines.
ACKNOWLEDGEMENTS
This work was supported by Seoul National University Bundang Hospital Research Fund no. 02-2007-008 (2007) and Seoul National University Hospital Grant no. 04-2008-0920 (2008).