Expression and Characterization of α-Methylacyl CoA Racemase from Anisakis simplex Larvae
Article information
Abstract
Larval excretory-secretory products of Anisakis simplex are known to cause allergic reactions in humans. A cDNA library of A. simplex 3rd-stage larvae (L3) was immunoscreened with polyclonal rabbit serum raised against A. simplex L3 excretory-secretory products to identify an antigen that elicits the immune response. One cDNA clone, designated as α-methylacyl CoA racemase (Amacr) contained a 1,412 bp cDNA transcript with a single open reading frame that encoded 418 amino acids. A. simplex Amacr showed a high degree of homology compared to Amacr orthologs from other species. Amacr mRNA was highly and constitutively expressed regardless of temperature (10-40℃) and time (24-48 hr). Immunohistochemical analysis revealed that Amacr was expressed mainly in the ventriculus of A. simplex larvae. The Amacr protein produced in large quantities from the ventriculus is probably responsible for many functions in the development and growth of A. simplex larvae.
Anisakis simplex (Nematoda: Anisakidae) is one of the most frequently encountered parasites in humans through the ingestion of marine products. A. simplex has a complicated life cycle that involves sea mammals, crustaceans, cephalopods, and fish. The human anisakiasis occurs when people eat raw or undercooked fish or squid. A. simplex can cause direct tissue damage following larval invasion in the gut wall, development of an eosinophilic granuloma, or perforation of the gut [1,2]. It has also been reported that an A. simplex larval infection can cause an allergic response due to anti-Anisakis IgE antibodies [3-5]. The allergic responses caused by anisakiasis include anaphylaxis, urticaria, angioedema, rheumatic manifestations, and nephrotic syndrome [6-11]. These allergic responses have been reported to be associated with A. simplex larvae excretory-secretory (ES) products. Immunoblot analysis of A. simplex ES products from the 3rd-stage larvae (L3) revealed several allergenic bands ranging from 10 to 186 kDa [12]. Moreover, the ES products from A. simplex larvae elicited proinflammatory cytokine and chemokine secretion from a mouse lung epithelial cell line and primary lung epithelial cells [13]. Although infected larvae are unable to complete their life cycle in the human body, they can survive a few weeks and even develop to further stages [14]. The human immune system can be activated with ES products during the developmental process of the larvae. It is usually assumed that there would be a plenty of ES products that could provoke immune responses in an infected human.
We immunoscreened cDNA library of A. simplex larvae with polyclonal rabbit serum raised against major ES products (35-40 kDa) of larvae to identify an antigen that elicits the immune response after A. simplex larval infection. This approach led to the identification of α-methylacyl CoA racemase (Amacr), which is an essential enzyme involved in the β-oxidation of bile acid intermediates and branched-chain fatty acid degradation. We also analyzed its homology with other Amacr orthologs, its regulation by RT-PCR, and localization by immunohistochemistry.
Live A. simplex larvae were collected from the body cavity and the viscera of the chub mackerel, Scomber japonicus, which were purchased from a local fish market. To obtain ES products, live L3 were incubated in DMEM (Dulbecco's Modified Eagle Medium, gentamycin 150 mg/ml, vancomycin 10 mg/ml, and 0.5 ml/larva) at 37℃ for 48 hr. The ES products were centrifuged and concentrated (Amicon stirred cells, Millipore Corp., Billeria, Massachusetts, USA). After the concentrated ES products were resolved by SDS-PAGE on a 12% gel, the strong protein bands between 35 and 40 kDa were cut out and minced with a razor blade (Fig. 1). The finely minced gel was mixed with Freund's adjuvant and PBS to make a homogeneous suspension. The suspension (1 ml) was injected intraperitoneally into 8 to 10 week-old ICR mice. After the last injection conducted at 1-week intervals, the same suspension was mixed with Freund's incomplete adjuvant and administered intraperitoneally. The sera were collected 2 weeks later.
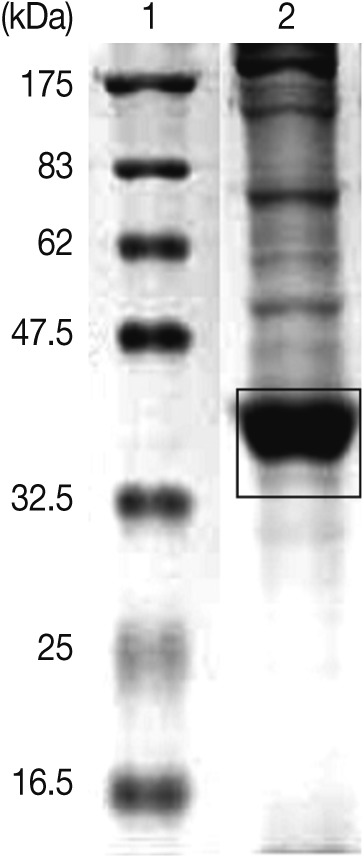
SDS-PAGE of excretory-secretory (ES) products from Anisakis simplex L3 at 37℃ for 24 hr. ES products ranging from 35 to 40 kDa were injected into mice for the production of polyclonal antibodies. Lane 1, molecular marker; Lane 2, ES products.
An A. simplex L3 lambda Zap II library [15] was screened using 1:500 diluted polyclonal anti-major Anisakis ES (against 35-40 kDa) mouse sera (pre-absorbed with Escherichia coli lysates) and 1:1,000 diluted horseradish peroxidase-labeled goat anti-mouse IgG (Serotec, Oxford, UK). Nylon transfer membranes (Amersham Biosciences, Piscataway, New Jersey, USA) were developed using DAB substrate (Thermo Fisher Scientific Inc., Pittsburgh, Philadelphia, USA). Positive plaques were picked and purified by plating at successively lower densities. Immunopositive plaques from the final round of purification were subcloned using the lambda ZAP II in vivo excision method (ExAssist/SOLR system; Stratagene, La Jolla, California, USA). One clone was isolated, and then the purified plasmid DNA was sequenced (Macrogen, DNA Sequencing Service, Seoul, Korea).
Nucleotide and amino acid sequences of the selected clone were compared via BLAST analysis at the National Center for Biotechnology Information. Twenty amino acid sequences, including the A. simplex Amacr, were chosen and analyzed. The significance and evolutionary conservation of Amacr was investigated by constructing a phylogram using the maximum Parsimony method [16] in MEGA4 [17]. Amino acid sequence accession numbers are as follows; A. simplex (ADW54022.1), Caenorhabditis elegans (NP_509517.2), Sus scrofa (XP_0031339 21.1), Danio rerio (NP_001018462.1), Gallus gallus (NP_00102 6619.1), Bos taurus (AAI18287.1), Anopheles gambiae (XP_3170 33.2), Xenopus laevis (NP_001167512.1), Ailuropoda melanoleuca (XP_002919481.1), Mus musculus (NP_032563.2), Branchiostoma floridae (XP_002595975.1), Homo sapiens (BAD96551.1), Canis familiaris (XP_855429.1), Taeniopygia gutata (XP_002191 692.1), Saccoglossus kowalevskii (XP_002730894.1), Oryctolagus cuniculus (XP_002714167.1), Equus caballus (XP_001500301.1), Drosophila melanogaster (NP_610054.1), Rattus norvegicus (NP_036948.1), and Pan troglodytes (XP_001152632.1).
The recombinant protein was expressed as a glutathione S-transferase (GST) fusion protein. The full-length sequence of the selected clone was subcloned into the pGEX-4T-2 vector (Amersham BioSciences) and was used to transform E. coli strain BL21. Bacteria were cultured in 1.0 mM IPTG from 35℃ to 37℃. The recombinant protein was purified using a MicroSpin GST Purification Module (Amersham Biosciences).
Western blotting was performed to check the reactivity of the recombinant protein obtained. The recombinant protein was separated by SDS-PAGE on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). Transferred proteins were confirmed by Ponceau staining and blocked with PBS containing 5% skim milk at room temperature for 1 hr. After washing 3 times with PBST for 10 min, membranes were incubated overnight with 1:500 rabbit polyclonal Amacr antibodies to AMCR (Abcam, Cambridge, UK) at 4℃. The membranes were then washed with TBST and incubated with 1:1,000 diluted horseradish peroxidase-labeled goat polyclonal antibodies to rabbit IgG (Abcam). The ECL plus Western blotting system (Amersham Biosciences) was used for detection.
To study the effects of incubation time and temperature on mRNA expression of the selected clone, 100 A. simplex larvae were incubated in DMEM at 10℃, 15℃, 25℃, 37℃, and 40℃ for 24 or 48 hr. After the total RNA was extracted (TRIzol, Invitrogen, Carlsbad, California, USA) and reverse transcribed using RT-PCR Master Mix (Takara, Tokyo, Japan), PCR was performed with Amacr primers (5'-AGC CCG AGG TAA AAC CAT TA and 3'-ACA ATC ACC CGA AAA CCA C). PCR cycles consisted of an initial 5 min denaturation at 94℃ and 30 cycles of 94℃ (30 sec), 56℃ (30 sec), 72℃ (30 sec), followed by a final 7 min extension at 72℃. The PCR products were analyzed by electrophoresis on a 2% agarose gel.
Immunohistochemistry was performed to confirm the distribution of the Amacr protein. Paraffin-embedded larvae slides were deparaffinized and hydrated. For antigen retrieval, slides were immersed in citrate buffer (0.01 M, pH 6.0) and heated twice in a microwave (700 W) for 5 min. The slides were then quenched with endogenous peroxidase by incubation with a 3% hydrogen peroxide solution for 5 min and washed 3 times in PBS for 5 min. Slides were immunostained with rabbit polyconal Amacr antibody (1:500 dilution, Abcam) at 4℃ overnight. After that slides were washed 3 times with PBS for 5 min and incubated with horseradish peroxidase-labeled goat polyclonal antibodies to rabbit IgG (1:2,000 dilution, Abcam) for 2 hr at room temperature. Slides were washed 4 times with PBS for 5 min each. The color reaction was developed with Dako's EnVision™ System (DAKO, Carpinteria, California, USA). Slides were stained with DAB and counterstained with Meyer's hematoxylin (DAKO) for 20 sec, dehydrated, and mounted with Permount (Fisher Scientific) and observed by microscopy (Nikon, Eclipse 80i, Star Capture Basic V1.0, Tokyo, Japan).
We selected 1 clone from the final round of immunoscreening, designated as Asim37, which was the most consistently reactive with the polyclonal anti-Anisakis ES mouse sera during the screening process. The Amacr clone had an insert size of 1,412 bp and yielded 1 open reading frame (ORF) that encoded for a 418 amino acid protein with a predicted molecular weight of 46.7 kDa (GenBank; HQ662605). The complete sequence of the Amacr clone was identified as a α-methylacyl CoA racemase ortholog, a member of the CoA-transferase family III (Fig. 2A). Sequence analysis showed that the cDNA included 5' (32 bp) and 3' (188 bp) untranslated sequences. The cDNA construct contained an ATG start codon, a TAA stop codon, and a poly-A tail that comprised of 31 adenine residues. A typical polyadenylation signal was also present in 13 nucleotides upstream of the poly-A sequence. Interestingly, the A. simplex Amacr amino acid sequence was longer than that of other species at the N-terminus. Sequence homology analysis of the Anisakis Amacr gene displayed a homology of 36-50% and highly conserved amino acid sequences compared with 19 Amacr orthologs of different species (data not shown). The sequence analysis and phylogenetic analysis classified A. simplex Amacr as the most closely related to the C. elegans group (Fig. 2B).
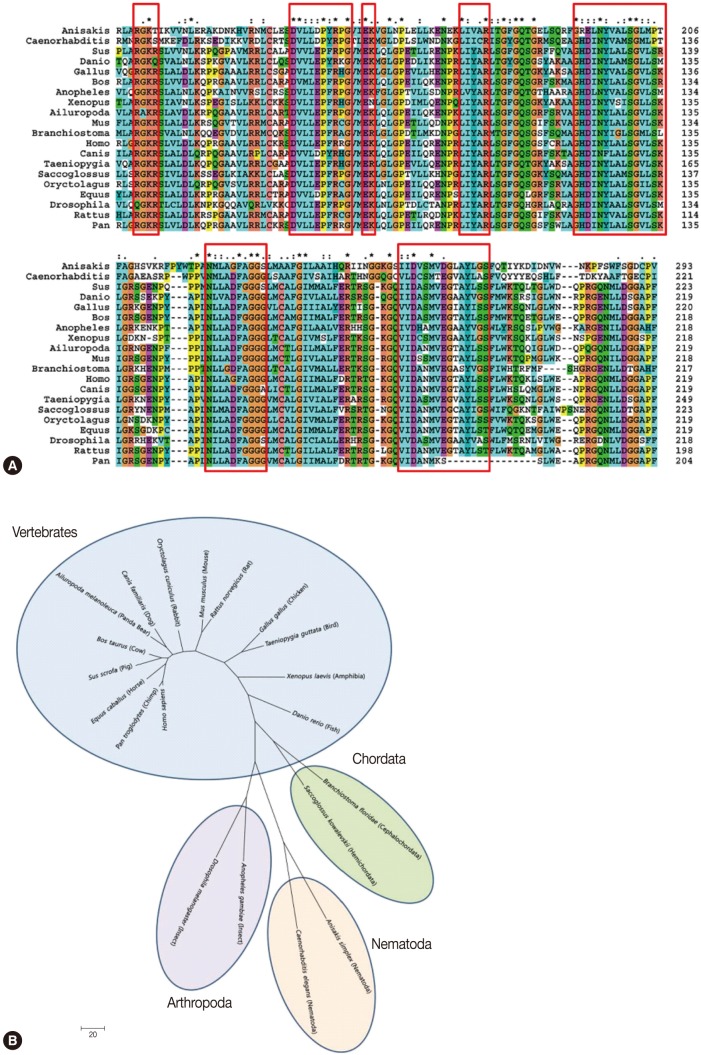
Complete nucleotide and amino acid sequences (A) and phylogenetic analysis (B) of α-methylacyl CoA racemase (Amacr) from Anisakis simplex L3 (GenBank; HQ662605). The Amacr gene consists of 1,414 bp and 418 amino acids. The start codon (ATG), stop codon (TAA), and poly-adenylation signal (aataaa) are marked with an asterisk. The phylogenetic analyses were conducted in MEGA4 [23] using the maximum Parsimony method. The A. simplex Amacr was grouped with the nematode Caenorhabditis elegans.
The expression of the Anisakis Amacr was constitutively high under all conditions tested, suggesting that a large amount of Amacr gene expression in A. simplex L3 is required over a long period for the larval survival (Fig. 3). The expression of recombinant A. simplex Amacr protein was confirmed with SDS-PAGE. The GST-fusion protein revealed a 73 kDa band on the gel (Fig. 4). The recombinant Amacr protein showed a reactive band against Amacr antibody, a rabbit polyclonal to AMCR (Abcam) (Fig. 5). Immunohistochemical analysis showed that the Amacr protein was mainly localized at the ventriculus of A. simplex larvae, regardless of different time points (24 and 48 hr) (Fig. 6).
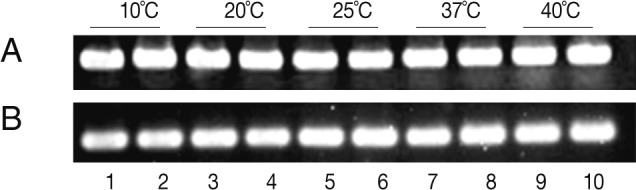
RT-PCR anaylsis of the Amacr expression in Anisakis simplex L3 according to time and temperature. (A) α-actin. (B) RT-PCR of the Amacr mRNA. Lane 1-2, 10℃; Lane 3-4, 20℃; Lane 5-6, 25℃; Lane 7-8, 37℃; Lane 9-10, 40℃. Lanes 1, 3, 5, 7, and 9, 24 hr incubation; Lanes 2, 4, 6, 8, and 10, 48 hr incubation. The Amacr expression level was high and constitutive, regardless of time and temperature.
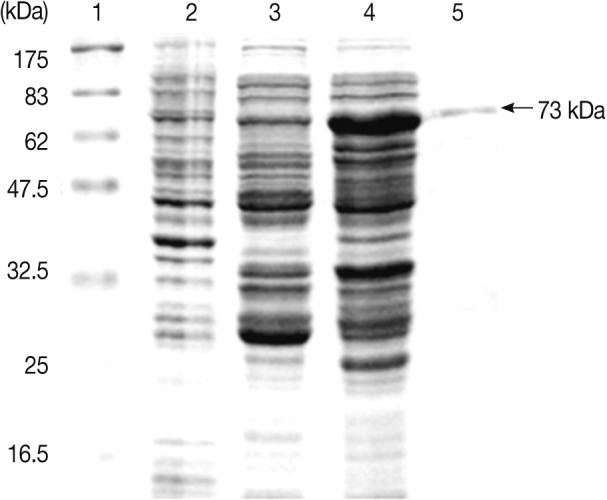
Expression of the Anisakis simplex L3 Amacr in E. coli. Lane 1, molecular marker; Lane 2, BL21(DE3); Lane 3, pGEX 4T-2; Lane 4, IPTG-induced; Lane 5, purified recombinant protein. The recombinant protein was constructed as a GST fusion protein and purified with a MicroSpin GST purification module. A GST fusion protein of the A. simplex Amacr was presented as 73 kDa band on the gel.
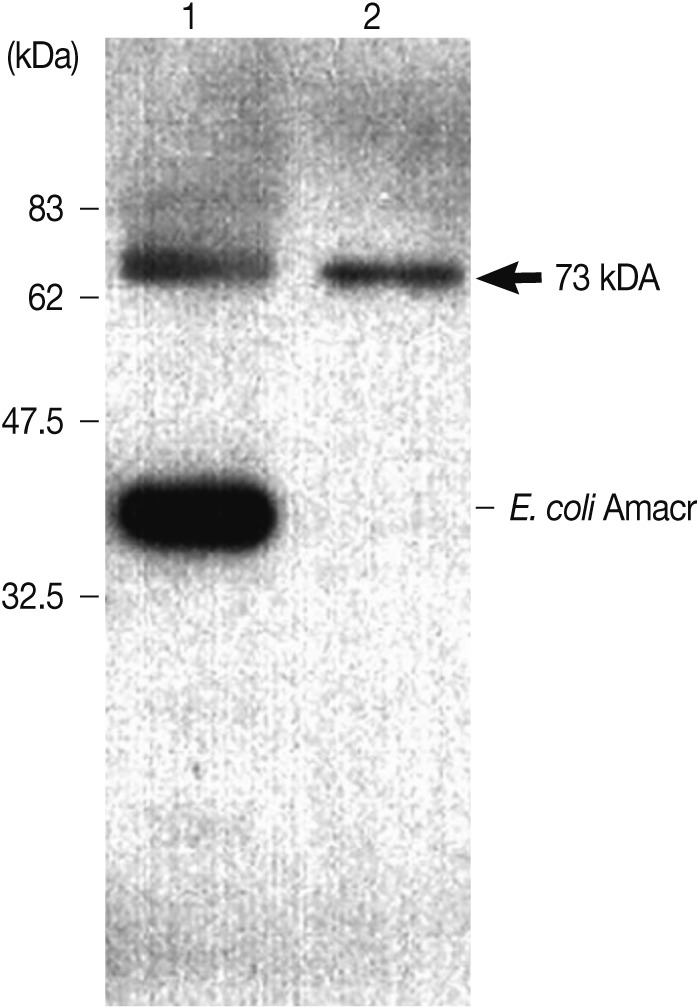
Immunoblot analysis of the recombinant Amacr protein with rabbit polyclonal Amacr antibodies. Lane 1, IPTG-induced BL21(DE3); Lane 2, purified recombinant protein Amacr-GST fusion protein marked by an arrow (73 kDa). The recombinant Amacr protein showed a single reactive band against the Amacr antibody, rabbit polyclonal to AMCR (Abcam).
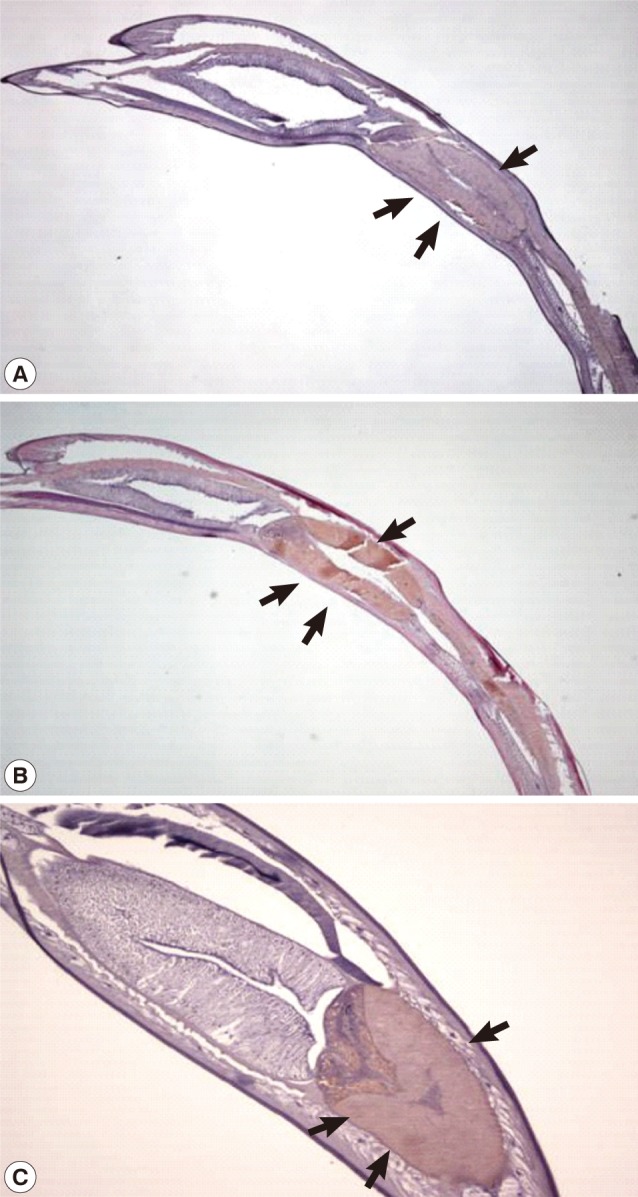
Expression of the Amacr in Anisakis simplex larvae. The Amacr product was observed primarily in the ventriculus. (A) Control staining of larval ventriculus (arrows) with normal mouse serum (×40). (B) Specifically stained ventriculus (arrows). The L3 were incubated at 24℃ for 24 hr (×40). (C) Larval Amacr immunostaining incubated for 48 hr also represents the same localization as 24 hr larvae. Ventriculus (arrows) (×100). Immunohistochemical study showed that the Amacr product was found specifically in the ventriculus of the larvae.
We have demonstrated the Amacr gene from A. simplex larvae. The Amacr gene contained a 1,412 bp cDNA transcript that encoded 418 amino acids. The expression of Amacr mRNA was constitutively high regardless of temperature (10-40℃) and time (24-48 hr). Amacr protein was expressed mainly in the ventriculus of A. simplex larva. The phylogenetic analysis showed that this gene was clustered to C. elegans which was the only nematode species that the Amacr gene has been known.
The Amacr gene encodes a racemase. The encoded enzyme racemase interconverts pristanoyl-CoA and C27-bile acyl CoAs between their (R)- and (S)-stereoisomers. The conversion to the (S)-stereoisomers is necessary for degradation of these substrates by peroxisomal β-oxidation. Encoded enzyme from this locus localizes at both mitochondria and peroxisomes. The function of this enzyme is presumed to be involved in energy production. In humans, an Amacr overexpression, deficiency, or mutant variants are known to be associated with a disease state [18,19]. Specifically, the Amacr overexpression is considered one of the most potent diagnostic markers of the prostate cancer [20].
It is interesting to note that the function of the Amacr is related to the synthesis of the bile acid. The bile acid has long been known to play an important role in the host-parasite relationships, including penetration, immune suppression, and inflammation or modulation of homeostasis, enabling parasite invasion, and establishment [21]. The Amacr gene from A. simplex is thus presumably involved in the parasite-host relationships.
Generation of ATP from lipids occurs within mitochondria via the β-oxidation of fatty acids. However, the β-oxidation of fatty acids has not been reported in adult or larval helminths due to their anaerobic condition within the host even though the genes for the TCA cycle are present in several nematodes [22,23]. Recently, the oxidization of fatty acids and the activity of fatty acid oxidation enzymes were confirmed in infective larvae of Angiostrongylus cantonensis [24]. The uptake of arachidonic acid by A. cantonensis and the presence of oxidation enzymes strongly suggest that some nematodes can use the β-oxidation pathway.
The Amacr mRNA expression did not change depending on the incubation time (24 and 48 hr) and temperature ranging from 10℃ to 40℃. These results imply that a large amount of the Amacr gene expression is required over a long period of time. The expression of high levels of the Amacr gene in A. simplex L3 can contribute to the generation of energy for further development. The immunohistochemical findings revealed that the Amacr was mainly localized in the ventriculus. The secretion from the ventriculus can play many roles, including invasion of the gastrointestinal mucosa of the hosts.
In conclusion, we have demonstrated the Amacr gene from A. simplex larval cDNA library by immunoscreening with ES antibody. Our data showed that A. simplex Amacr needs a large amount over an extended period of time to involve in energy production for the larval development.
ACKNOWLEDGMENT
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2006-E00037).