Keratitis by Acanthamoeba triangularis: Report of Cases and Characterization of Isolates
Article information
Abstract
Three Acanthamoeba isolates (KA/E9, KA/E17, and KA/E23) from patients with keratitis were identified as Acanthamoeba triangularis by analysis of their molecular characteristics, a species not previously recognized to be a corneal pathogen. Epidemiologic significance of A. triangularis as a keratopathogen in Korea has been discussed. Morphologic features of Acanthamoeba cysts were examined under a microscope with differential interference contrast (DIC) optics. Mitochondrial DNA (mtDNA) of the ocular isolates KA/E9, KA/E17, and KA/E23 were digested with restriction enzymes, and the restriction patterns were compared with those of reference strains. Complete nuclear 18S and mitochondrial (mt) 16S rDNA sequences were subjected to phylogenetic analysis and species identification. mtDNA RFLP of 3 isolates showed very similar patterns to those of SH621, the type strain of A. triangularis. 16S and 18S rDNA sequence analysis confirmed 3 isolates to be A. triangularis. 18S rDNA sequence differences of the isolates were 1.3% to 1.6% and those of 16S rDNA, 0.4% to 0.9% from A. triangularis SH621. To the best of our knowledge, this is the first report, confirmed by 18S and 16S rDNA sequence analysis, of keratitis caused by A. triangularis of which the type strain was isolated from human feces. Six isolates of A. triangularis had been reported from contaminated contact lens cases in south eastern Korea.
INTRODUCTION
Acanthamoeba keratitis (AK) is a serious corneal infection caused by the genus Acanthamoeba which is abundant in soil, air, fresh water, ocean sediment [1]. AK has become exponentially increased in recent years correlating to the growing number of contact lens wearers [2,3]. Currently, 8 species of Acanthamoeba, i.e., A. castellanii, A. polyphaga, A. rhysodes, A. culbertsoni, A. hatchetti, A. griffini, A. quina and A. lugdunensis, have been identified in ocular infection [4-6].
Identification of Acanthamoeba isolates at the species level has been tried by several methods. Pussard and Pons classified Acanthamoeba into 3 groups according to the size and morphological features of the amebic cyst [7]. However, because of variable cyst morphology by culture conditions, species identification by morphology alone can hardly be possible. Alloenzyme analysis and mitochondrial DNA restriction fragment length polymorphism (mtDNA RFLP) studies have also been widely applied, but the results were too pleomorphic to be the criteria for species identification [8,9]. Recently nuclear 18S and mt 16S rDNA sequence analyses has been reported particularly useful in taxonomic and epidemiological studies of Acanthamoeba spp [10-13].
In the present study, we describe the laboratory investigation of 3 ocular isolates (KA/E9, KA/E17, and KA/E23) that were finally identified as A. triangularis by mtDNA RFLP and nuclear 18S and mt 16S rDNA sequence analyses.
MATERIALS AND METHODS
Case records
Case 1
A 32-yr-old male, daily-wear soft contact lens (CL) user developed ocular pain, tearing and conjunctival injection on his right eye after taking off contact lens. At first, he was diagnosed and treated as CL-induced bacterial kerititis with fortified cephalosporin and gentamicin for 1 month. However, his keratitis became worse with treatment, progressed to corneal ulcer and he was referred for diagnosis and proper treatment. On the first examination of the affected eye, there was a round central corneal ulcer with elevated necrotic corneal tissue. Surrounding corneal tissue was moderately infiltrated and endothelial cell deposits existed. Anterior chamber reaction was moderate degree (Fig. 1A).
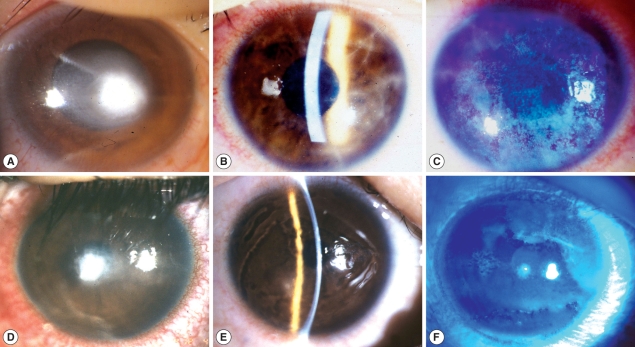
Clinical appearance of infected corneas of 3 cases. (A) Right eye of case 1; (B, C) Right eye of case 2; (D) Right eye of case 3; (E,F) Left eye of case 3; B & E: Slit lamp examination.
Visual acuity of right eye was FC/30 cm. With the suspicion of mixed infection including Acanthamoeba, treatment was begun with fortified antibiotics, topical polyhexamethylene biguanide (PHMB) (0.02%) and systemic itraconazole. Cultures of corneal scrapings for the bacteria, fungi and Acanthamoeba were done. Other cultures except for Acanthamoeba were negative. Three months after antiamebic treatment, corneal ulcer was healed with residual corneal opacity. Visual acuity improved to 20/200 with glasses correction.
Case 2
A 17-yr-old female was presented with ocular pain, photophobia and vision decrease on her right eye after using disposable soft contact lens. She had used contact lens intermittently for 1 yr, and stored contact lens in contact lens case while not using. The corrected vision of her right eye was 20/50. On slit-lamp examination, there was diffuse stromal infiltrates with typical perineuritis over the whole cornea (Fig. 1B). The corneal epithelial layer was haze due to diffuse erosion and irregularity. The conjunctiva was moderately injected (Fig. 1C). She was clinically diagnosed as AK and treated with antiamebic agent, PHMB 0.02%, and systemic itraconazole. Culture of conjunctival swab and solution of contact lens case revealed Acanthamoeba. After 3-month antiamebic treatment, the corrected vision of her right eye improved to 20/25. The cornea healed with residual irregular subepithelial opacity. Other ocular findings were within normal range.
Case 3
A 16-yr-old female was referred for the diagnosis of suspicious Acanthamoeba infection. Patient had used orthokeratology lens for 2 yr for the correction of myopia. While using the orthokeratology lens, she experienced ocular pain, tearing and decreased vision in both eyes, specially left eye. She was treated for CL-induced infectious keratitis for 3 wk but ocular symptoms more aggravated. On the first examination, her corrected vision was 20/25 in right eye and FC/50 cm in left eye. On slit-lamp examination, epithelial surface irregularity, stromal infitrates and peripheral polyneuritis were shown in right eye (Fig. 1D). There was a 2 mm-size central corneal ulcer with stromal infiltrates in left eye (Fig. 1E, F). With clinical diagnosis of acanthamoebic infection, topical PHMB (0.02%) and systemic itraconazole were started. Culture for Acanthamoeba was positive from corneal scarping on left eye. Two months after antiamebic treatment, visual acuity of right eye recovered to 20/20 and 20/25 in left eye with glasses correction. Right cornea was clear except paracentral dot opacity but left one healed with faint diffuse central stromal opacity.
Protozoology
The corneal epithelial scrapings from patients were obtained by using sterile scalpels, and conjunctival swab by sterile cotton ball. The samples were inoculated onto 1.5% agar plates covered with heat-treated Escherichia coli (ATCC 25922, Washington D.C., USA). The plates were incubated at 25℃ and examined daily for the presence of Acanthamoeba under an inverted microscope. Single encysted ameba was transferred into a new agarcoated plate and some cysts from each plate were subjected to axenization. The isolates were designated Korean Acanthamoeba/Eye series KA/E9, KA/E17, and KA/E23.
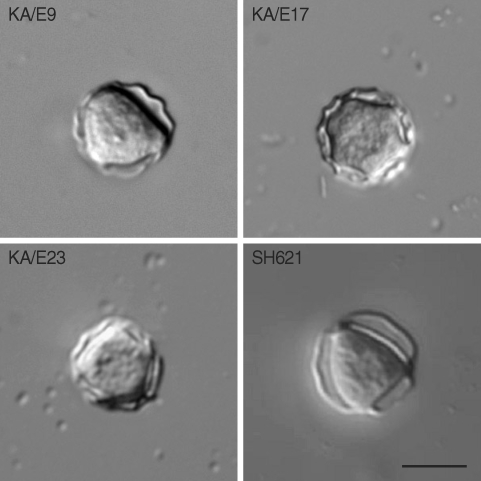
A piece of agar plate covered with many cysts of a clone was treated with 0.1 N HCl for 24-36 hr for axenization and washed with PBS 3 times. The piece of agar plate was placed in Peptone-Yeast-Glucose (PYG) medium at 25℃ incubator (Sanyo, San Diego, California, USA). The size and number of arms of the cysts were measured under a microscope with DIC optics.
Mitochondrial (mt) DNA restriction fragment length polymorphism (RFLP)
The Acanthamoeba mtDNA was extracted via the method described by Yagita and Endo [14]. Acanthamoeba trophozoites harvested at the end of the logarithmic growth phase were washed in cold PBS. The amoebas were lysed with fresh 1% sodium dodecyl sulfate (SDS) solution in 0.2 N NaOH and potassium acetate buffer, and incubated for 15 min on ice. The mtDNA was then extracted with phenol and phenol-chloroform (1 : 1) and recovered via precipitation with cold absolute ethanol in the presence of sodium acetate. The extracted mtDNA was then digested with the Eco RI, Bgl II, and Cla I restriction enzymes (Promega, Madison, Wisconsin, USA) at 37℃ overnight. The mtDNA RFLP patterns were evaluated via 0.7% agarose gel electrophoresis and photographed under a Gel Doc™XR 170-8171 (Bio-Rad, California, USA).
Sequence analysis of the mtDNA 16S rDNA and the 18S rDNA of Acanthamoeba
In order to amplify the mtDNA 16S rDNA of the Acanthamoeba, the mtDNA of each Acanthamoeba was employed as a template. The nucleotide sequences of the primers were as follows: forward; Mt R 5'-TTGGGTTTTATTCTGGCTCCGAGTGAACG-3'; Mt F 5'-AATTTTGTCCAGCAGCAGGTTCCC-3'. Mt 16S rDNA amplification reactions were separately conducted using a premix of 20 µl scale (iNtRON, Seongnam, Korea), which included 2.5 U of i-Taq™ DNA polymerase (iNtRON). The PCR reactions were conducted at an initial 2 min at 94℃, followed by 30 cycles of 94℃ for 30 sec, 60℃ for 30 sec, 72℃ for 2 min, and a final 5 min elongation step at 72℃.
The genomic DNA of Acanthamoeba isolates acquired via the method described by Kong and Chung was utilized as a template for 18S rDNA PCR [15]. The amplification of amebic 18S rDNA gene fragments was conducted using primers originating from the small subunit ribosomal RNA coding DNA (ssu rRNA) sequence of the A. castellanii Neff strain, as reported by Gunderson and Sogin [16]. The sequences of the primers were as follows: forward; 5'-CCGAATTCGTCGACAACCTGGTTGATCCTGCCAGT-3' reverse; 5'-GGATCCAAGCTTGATCCTTCTGCAGGTTCACCTAC-3'. The 18S rDNA amplification reactions were separately conducted using premix of 20 µl scale (iNtRON). The PCR reactions were conducted for an initial 3 min at 94℃, followed by 30 cycles of 94℃ for 1 min, 58℃ for 1 min, 72℃ for 2.5 min, and a final elongation step of 7 min at 72℃. The amplified PCR products were ligated into the pGEM-T easy cloning vector (Promega, Madison, Wisconsin, USA), with subsequent transformation into E. coli. DNA sequencing was conducted by an outside company (Macrogen, Seoul, Korea). Sequence data were deposited at GenBank and are available under the following accession numbers: AF316546 (KA/E9, 18S rDNA), EU515181 (KA/E9, 16S rDNA), AY148963 (KA/E17, 18S rDNA), EU572722 (KA/E17, 16S rDNA), EF140625 (KA/E23, 18S rDNA), and EU515182 (KA/E23, 16S rDNA).
Phylogenetic analyses
Nucleotide sequences were compared to the sequences of published strains (Table 1) via BLAST search [17]. Clustal X was used for pairwise alignment and calculations of the percentages of sequence dissimilarity [18]. Phylogenetic analysis was conducted using the Clustal X 1.82 program with a low gap penalty, and the phylogenetic trees were drawn using Tree View computer software.
RESULTS
Morphology of Acanthamoeba isolates
Cysts of Acanthamoeba isolates KA/E9, KA/E17, KA/E23 and A. triangularis exhibited typical morphologic characteristics of group II classified by Pussard and Pons [7]. They exhibited double walled cyst morphology consisting of thick wrinkled ectocysts and satellite or polygonal endocyst (Fig. 2). The cyst diameter varied 12.0-22.5 µm and the number of arms 3-6.
mtDNA RFLP
Three ocular isolates showed very similar mtDNA RFLP patterns with A. triangularis SH621 but quite different from A. castellanii Castellani by 3 kinds of restriction enzymes (Eco RI, Bgl II, and Cla I). mtDNA RFLP of KA/E9 and KA/E23 also showed extra bands (arrowheads) in additions to common fragments (Fig. 3).
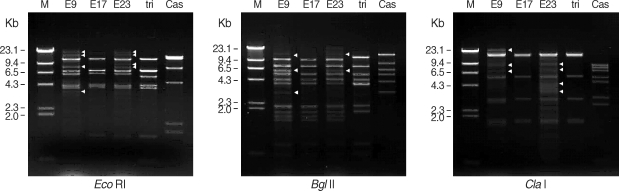
Agarose gel electrophoretic restriction fragment patterns of mitochondrial DNA of Acanthamoeba sp. KA/E9, KA/E17, KA/E23 and the reference strains by 3 kinds of restriction enzymes. M; Hind III digested λ phage DNA as DNA molecular size standard. Acanthamoeba KA/E9 and KA/E23 show extra bands (arrowheads) as well as common bands with KA/E17 and A. triangularis SH621. Cas is A. castellanii Castellani.
Nuclear 18S rDNA and mt 16S rDNA sequence analysis
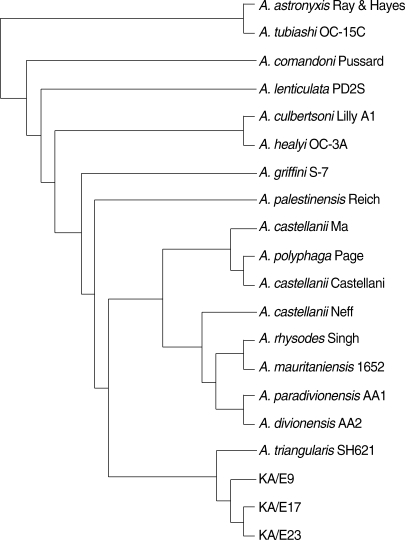
Pairwise sequence distances of nuclear 18S and mt 16S rDNAs among isolates are shown in Table 2. Nuclear 18S rDNA seuquences of Acanthamoebae KA/E9, KA/E17, and KA/E23 were compared with those of other Acanthamoeba reference strains stored in GenBank. They showed the most pronounced sequence similarity with A. triangularis SH621 (alignment data not shown). In a phylogenetic tree based on 18S rDNA sequences (Fig. 4), Acanthamoeba KA/E9, KA/E17, and KA/E23 were positioned to sequence genotype T4, the genotype encompassing the majority of clinical and environmental isolates of Acanthamoeba.

Divergence of the 18S rDNA and mt 16S rDNA sequences for isolates KA/E9, KA/E17, KA/E23 and reference strains
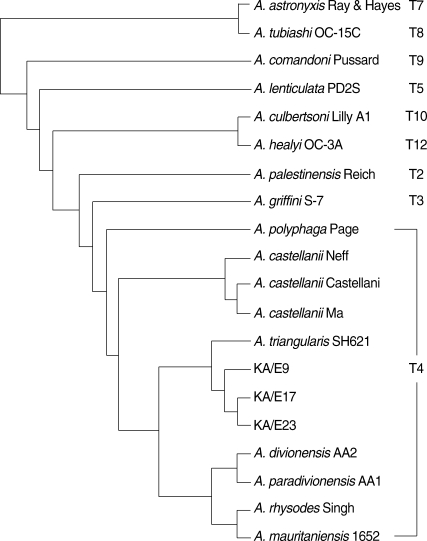
18S rDNA-based phylogenetic tree reflecting the affiliation of Acanthamoeba sp. KA/E9, KA/E17, and KA/E23 with selected members of genus Acanthamoeba.
In comparison of mt 16S rDNA sequences, they also showed a high degree of sequence similarity with A. triangularis SH621 (99.1% and 99.6%, respectively). In a phylogenetic tree based on 16S rDNA sequences (Fig. 5), 3 ocular isolates belonged to the same clade with SH621. In the light of the sequence homology of 18S and 16S rDNA with SH621, the type strain of A. triangularis, and the pattern of mtDNA RFLP, these 3 clinical isolates were identified as A. triangularis.
DISCUSSION
AK is severe, progressive and sight threatening infection of cornea. Since the first ocular infection with Acanthamoeba [3,19,20], the occurrence of AK has been escalating in correlation with the increasing number of contact lens wearers. Until now, 8 Acanthamoeba species have been reported to be able to cause keratitis in the literature: A. castellanii, A. polyphaga, A. rhysodes, A. culbertsoni, A. hatchetti, A. griffini, A. quina, and A. lugdunensis [5,6,21,22]. Three isolates KA/E9, KA/E17, and KA/E23 of the present cases showed 0.5% to 0.9% sequence difference of 18S rDNA to one another and 1.3% to 1.6% from SH621 strain, the type strain of A. triangularis. Both isolates also showed 0.3% to 0.7% sequence difference of 16S rDNA to one another and 0.4% to 0.9% from SH621 strain. These results suggest that the species of Acanthamoeba involved is identical to A. triangularis. This is the first report of AK caused by A. triangularis that was characterized and identified by mtDNA RFLP and nuclear 18S and mt 16S rDNA sequence analyses.
Contamination of contact lens care systems are usually regarded as the first step in the pathogenesis of AK in lens wearers [9,21,23]. A. triangularis, originally isolated from the human feces in France [7], had not been reported to be a keratopathogen. Among 3 surveys for Acanthamoeba contamination in contact lens care system in Korea [9,23,24], A. triangularis was demonstrated only in the survey in southeastern Korea. Six out of 28 isolates (21.4%) were identified as A. triangularis based on riboprints, mtDNA RFLP, and alloenzyme analysis. Two of these patients were residents in southeasten Korea (one in Daegu and the other in Busan). Acanthamoeba lugdunensis is one of the most frequently isolated species from AK in Korea (unpublished data), which was reported the most frequently detected amebic contaminant of contact lens cases throughout Korea. Previous report on A. lugdunensis [5] and the present report coincided well with amebic contamination pattern of contact lens care system. Considering possible difference in sensitivity of amoebae isolates to amebicidal drugs [25], periodic survey on contamination patterns of contact lens cases by Acanthamoeba and drug sensitivity assay according to individual isolates or strains should be carried out for successful medical cure.
The cyst morphology of these ocular isolates was overlapped with that of various species within the morphologic group II, especially sequence type T4. Jongwutiwes et al. [26] reported heterogenic cyst morphology within cysts from the same AK patients. Two isolates of the second case by Jongwutiwes et al., which they regarded coinfection by A. castellanii and A. triangularis, showed the same mtDNA RFLP pattern, but they did not compare the pattern with that of SH621. Although the possibility of the case caused by A. triangularis cannot be ruled out, keratopathogenicity of A. triangularis has not been confirmed until now. Comparison of molecular data of clinical isolates with type strains of various species should be done for the definitive identification [5,27].
Although speciation of the Acanthamoeba isolate is unlikely to be essential in such treatment of the associated keratitis, genetic characterization is necessary at least for molecular epidemiology [5,6]. mtDNA RFLPs is a potent technique for differentiating various strains within a species, but not appropriate tool for species identification of Acanthamoeba because of profound variation within a species [28]. Extra bands on the mtDNA RFLP of KA/E9 and KA/E23 were confirmed due to intracellular endosymbiotic bacteria by orcein staining and electron microscopy [29]. Effects of endosymbionts on virulence of Acanthamoeba are now being evaluated. Results from 18S rDNA sequence analysis coincided better with previous species assignment, compared with those from 16S rDNA, but availability of 16S rDNA sequence anaylsis should not be underestimated [11]. Schuster and Visvesvara [30] reported the vast majority of karatitis-causing strains belonged to T4 among 15 sequence types of 18S rDNA. The isolates of the present cases and A. triangularis SH621 also belonged to sequence type T4. Possibility of all the species within T4 to be keratopathogens should be evaluated. Otherwise, pathogenic and nonpathogenic strains may compose a species of sequence type T4. We would rather propose sequence type T4 including type strain of the type species of the genus, as A. castellanii species complex [9,10,27]. Furthermore, taxonomic validity of some species belonging to T4, for example A. paradivionensis, A. divionensis, and A. mauritaniensis, should be reevaluated [31].
All of 3 patients we report here were lens wearers. Two patients had been daily wearing soft contact lenses for several years and the other had worn orthokeratology contact lenses for approximately 1.5 yr. Number of young people wearing contact lenses has been increasing in Korea for correction of myopia and even for cosmetic purpose [32,33]. More attention should be paid to hygienic maintenance of contact lenses care system for prevention of AK [34].