A Case of Plasmodium ovale Malaria Imported from West Africa
Article information
Abstract
Malaria is a parasitic infection caused by Plasmodium species. Most of the imported malaria in Korea are due to Plasmodium vivax and Plasmodium falciparum, and Plasmodium ovale infections are very rare. Here, we report a case of a 24-year-old American woman who acquired P. ovale while staying in Ghana, West Africa for 5 months in 2010. The patient was diagnosed with P. ovale malaria based on a Wright-Giemsa stained peripheral blood smear, Plasmodium genus-specific real-time PCR, Plasmodium species-specific nested PCR, and sequencing targeting 18S rRNA gene. The strain identified had a very long incubation period of 19-24 months. Blood donors who have malaria with a very long incubation period could be a potential danger for propagating malaria. Therefore, we should identify imported P. ovale infections not only by morphological findings but also by molecular methods for preventing propagation and appropriate treatment.
INTRODUCTION
Malaria is a parasitic infection that infects 300-500 million people per year with Plasmodium spp., and about 2 million people die from the infection. The parasites are transmitted by female Anopheles spp. mosquitoes. When the mosquito takes blood, sporozoites are discharged into the bite site. Then, the sporozoites are carried to the hepatocytes. The sporozoites multiply rapidly, and many merozoites are produced. These merozoites are released into blood where they invade RBCs [1]. Four common species that infect humans are P. falciparum, P. vivax, P. malariae, and P. ovale [2]. The most common species in Korea is P. vivax, which has been a representative indigenous species for the past several centuries, although the prevalence was low [3]. However, the prevalence rapidly increased during the Korean War, which presented the disease to the world [4]. Recently, the possibility of infection by imported malaria has increased because of the increased number of people traveling to tropical areas. Two cases of imported P. ovale have been reported in Korea both from West Africa [5,6].
Here we present a third case of P. ovale imported from West Africa, which was diagnosed by a peripheral blood smear, real-time PCR, nested PCR, and sequencing analysis. Interestingly, the strain of the present P. ovale had a very long incubation period, which is the reason for reporting it.
CASE RECORD
Clinical history and laboratory findings
A 24-year-old woman visited the emergency room with fever and chills. The patient was an American without underlying diseases and had lived in Korea for about 6 months. She had traveled to Jeonju, Gyeongju, and Suwon, but did not visit the northern part of Gyeonggi-do or Gangwon-do, which are endemic areas of malaria in Korea. However, she stayed in Ghana, West Africa for about 5 months from August to December 2010. At that time, she did not have any symptoms related to malaria and did not receive any treatment for malaria. In addition, she did not travel to any other malaria endemic areas after that. The complete blood cell count showed slight leukopenia (WBC 3,240/µl), slight thrombocytopenia (platelet 138,000/µl) and hemoglobin 13.2g/dl. Biochemical tests (total protein, albumin, glucose, total bilirubin, aspartate aminotransferase, alanine aminotransferase, creatine phosphokinase, amylase, urea nitrogen, creatinine, sodium, potassium, and chloride) were all normal, but lactate dehydrogenase (LDH) increased slightly (322 U/L; reference range 106-230 U/L).
The rapid diagnostic test (RDT) for malarial antigen (SD Bioline Malaria antigen P.f/Pan rapid kit, Standard Diagnostics, Inc., Yongin, Korea) at the first visit was negative, and the fever had subsided, so the patient was discharged. However, on the next day, malarial parasites were found in the Wright-Giemsa stained peripheral blood smear. The patient was told to visit the hospital, but she did not return until she developed fever again 2 days later.
The Wright-Giemsa stained peripheral blood smear showed that the RBCs infected with malarial parasites were slightly enlarged and had fimbriated margins, which was suspicious of P. ovale (Fig. 1). After counting the number of malarial parasites per 200 WBCs using a microscope at ×1,000, the density of malarial parasites was calculated indirectly by counting the number of malarial parasites per µl using the number of WBCs measured by an automated hematology analyzer (XE-2100 hematology analyzer, Sysmex, Kobe, Japan). The densities of malarial parasites for the first and second visit were 583/µl and 2,478/µl, respectively.

Wright-Giemsa stained peripheral blood smear at the time of presentation (×1,000) shows different stages of Plasmodium ovale. Young and old ring-shaped trophozoites (A, B), older ameboid trophozoites (C), and schizonts (D) in enlarged red blood cells with a fimbriated margin are observed.
The patient was treated with hydroxychloroquine sulfate (immediate dose of 800 mg, followed by 400 mg at 6, 12, 18, and 24 hr) and primaquine (15 mg/day for 2 weeks). Her symptoms began to subside after 2 days of treatment, and no malarial parasites were observed in the peripheral blood on day 5 of treatment. On the 10th day of treatment, the complete blood cell count and biochemical test returned to normal, and malarial parasites were not observed.
Plasmodium genus-specific real-time PCR
Plasmodium genus-specific real-time PCR was performed with TaqMan fluorogenic probes targeting 18S rRNA (small subunit ribosomal RNA) genes to evaluate the presence of Plasmodium. The EDTA-anticoagulated blood specimen from the second visit was used. The AccuPower Malaria Quantitative PCR kit (Bioneer Co., Daejeon, Korea) and Exicycler™ 96 Real-Time Quantitative Thermal Block (Bioneer) for the thermal cycler were used. The PCR reagent mixture contained the Plasmodium genus-specific primer set, dual-labeled fluorogenic probes, DNA polymerase, dNTPs, and stabilizers. DNA was extracted with a DNA extraction kit from the same manufacturer (AccuPrep genomic DNA extraction kit, Bioneer). Real-time PCR was performed according to the manufacturer's instructions. The experiment included an internal positive control (IPC), 5 different concentrations of standards, a negative control (reaction mixture without DNA), and the patient's specimen, which was run in duplicate. The cycle threshold (Ct) for a positive result was <40. ExiDiagnosis analysis software (Bioneer) was used for the data analysis. Amplification curves of the 5 standards and IPC were determined to be valid. The patient's specimens showed Ct values of 25.7 and 25.5, indicating positive for Plasmodium spp. (Fig. 2).
Plasmodium species-specific nested PCR
Nested PCR targeting 18S rRNA genes was performed with primers and amplification conditions described by Snounou et al. [7] to identify Plasmodium spp. DNA was extracted from EDTA-anticoagulated blood samples with the QIAamp DNA Mini kit (QIAGEN Inc., Chatsworth, California, USA). The Plasmodium genus-specific primer set (rPLU5/rPLU6) was used for the first-round PCR. A second-round PCR was performed to identify the parasitic species present in the patient's sample. Four species-specific primer sets (P. falciparum, rFAL1/rFAL2; P. vivax, rVIV1/rVIV2; P. malariae, rMAL1/rMAL2; P. ovale, rOVA1/rOVA2) were used in the second-round PCR. Nested PCR was performed using a DNA thermal cycler (iCycler Thermal Cycler, Bio-Rad Laboratories, Hercules, California, USA). Thermal cycling was carried out under the following conditions: initial denaturation at 95℃ for 5 min, annealing at 58℃ for 2 min, and extension at 72℃ for 2 min (25 cycles); denaturation at 94℃ for 1 min, final annealing at 58℃ for 2 min, and final extension at 72℃ for 5 min (30 cycles). The 4 previously diagnosed clinical samples (P. falciparum, P. vivax, P. malariae, and P. ovale) and sterile distilled water were used as the positive and negative controls, respectively. The amplified DNA products corresponded to Plasmodium, 1,100 bp; P. falciparum, 205 bp; P. vivax, 120 bp; P. malariae, 144 bp; and P. ovale, 800 bp (Fig. 3). The presence of P. ovale in the patient's specimen was confirmed by nested PCR.
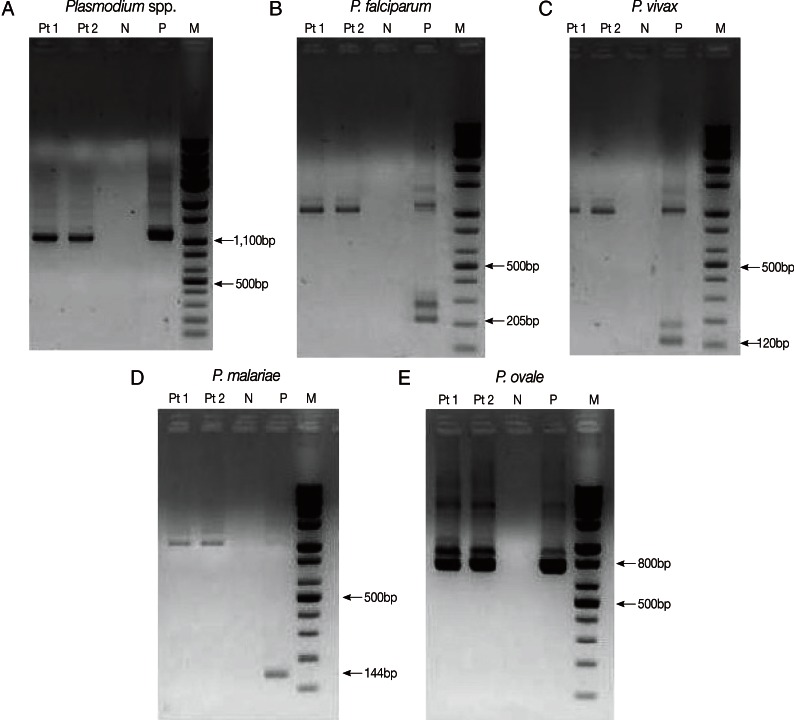
Plasmodium species-specific nested PCR targeting 18S rRNA. Plasmodium spp. (A) was identified by the first-round amplification (1,100 bp). P. falciparum (B), P. vivax (C), P. malariae (D), and P. ovale (E) were identified by second-round amplification. The 205 bp, 120 bp, 144 bp, and 800 bp fragments correspond to P. falciparum (B), P. vivax (C), P. malariae (D), and P. ovale (E), respectively. P. ovale was identified from the patient's sample (E). Pt1, Pt2, patient's samples in duplicate; N, negative control; P, positive sample; M, DNA size marker.
Sequencing analysis
The amplification products acquired from the second PCR (800 bp) were extracted from the agarose gel to perform sequencing analysis. The primer sets used in the nested PCR were also used, and the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, California, USA) and ABI 3730 XL autoanalyzer (Applied Biosystems) were used. The sequence alignment was performed using the single-stranded rRNA sequences in GenBank (P. ovale A-type, accession no. L48987) [8]. The isolated strain showed 99% similarity with P. ovale.
DISCUSSION
Malaria is a parasitic disease transmitted by mosquitoes and is characterized by periodic and convulsive high fever and chills. P. vivax malaria is the most common species and has been an indigenous infectious disease in Korea for several centuries [3]. However, the prevalence of malaria has decreased due to malaria control projects in the 1960s. Since 1984, there had been no reports of occurrence; however, the incidence began to increase from 1993 mainly among people in the military in the northern part of Gyeonggi-do [9]. Occasional reports of imported P. falciparum malaria have been made because of the increased number of overseas trips and overseas workers [10].
Among the 4 species, P. ovale malaria is generally confined to tropical West Africa, such as Nigeria and Ghana, as well as the Middle East, Papua New Guinea, and Indonesia. However, P. ovale malaria has also been reported from Southeast Asia, such as Myanmar, Cambodia, and Vietnam [11,12]. The incubation period of P. ovale is 10-17 days [11]. The dormant P. ovale hypnozoites remain quiescent in the liver and multiply later, which results in the prolonged incubation period or reccurrence [2]. Han et al. [5] reported a case of P. ovale infection imported from West Africa. The patient suffered from malaria of an unknown species while he stayed in West Africa and then returned to Korea. The patient developed symptoms of malaria 9 months after his return and was diagnosed with P. ovale infection. In the case report of Moon et al. [6], the patient stayed in West Africa for 20 days and he was diagnosed with P. ovale infection 3 months after he came back to Korea. However, the patient in our case was an American who had stayed in Korea for 6 months. Based on her travel history, real-time PCR, nested PCR, and sequencing analysis, the patient was considered infected during her stay in Ghana in 2010 rather than in Korea. Then, the incubation period would be 19-24 months.
P. ovale infection is generally less severe with less recurrence and good recovery. Because P. ovale invades only the reticulocytes, parasitemia is limited to 2-5% of RBCs [13]. The patient showed slight leukopenia and slight thrombocytopenia, and biochemical tests were all normal except LDH. After starting the hydroxychloroquine treatment, the complete blood cell counts and biochemical tests all returned to normal and the patient successfully recovered without any complications.
The rapid diagnostic test (RDT) for malarial antigen can quickly identify the presence of malarial infection by measuring parasite lactate dehydrogenase (pLDH). However, the RDT was negative in our case, even though malarial parasites were observed in a peripheral blood smear. According to the study by Park et al. [14], low density parasitemia can be the reason for a false-negative result. However, the parasite densities in our case were 583/µl and 2,478/µl for the first and second hospital visits, respectively. In addition, real-time PCR showed that there was 4.67×106-5.30×106 copies/ml as calibrated by the standard curves, which is not a low density parasitemia. Park et al. [12] also mentioned that the RDT could show a false-negative result due to the prozone effect with a very high density of >2,000 parasites/µl. Nevertheless, the patient's specimens from the first and second hospital visits were all negative on the RDT, which cannot be explained by the prozone effect. This strain might produce a very small amount of pLDH, or there might be a mutation in the gene that encodes pLDH. Further investigations are needed.
According to the exclusion criteria for blood donation used at the Korea Center for Disease Control and Prevention (issued in June 2011), no one can donate whole blood or platelet concentrate for 3 years if they have lived or have been in the military for 6 months or more in a high risk malarial infection area. If they traveled to an endemic area for 1 day to less than 6 months, they cannot donate whole blood or platelet concentrate for 1 year. In our case, the patient had lived in a malarial endemic area for 5 months. Thus, she could be a candidate for blood donation 1 year after returning from Ghana. This would be a potential source of malarial propagation through transfusion. Therefore, it is would be necessary to consider including molecular methods for malaria in screening tests for blood donors who have traveled to endemic areas, regardless of the period.
In our case, the patient stayed in Ghana for 5 months about 2 years ago and was negative on the RDT; however, malarial parasites were observed in the peripheral blood smear. She was confirmed to have P. ovale infection based on Plasmodium genus-specific real-time PCR, Plasmodium species-specific nested PCR, and sequencing analysis. Considering the test results and her travel history, we think that she was infected with P. ovale during her stay in Ghana rather than in Korea. In particular, she was infected with a strain with a very long incubation period of 19-24 months.
Therefore, careful history taking, including the current residency and travel history, is important to identify Plasmodium spp. accurately for appropriate treatment. Introducing molecular methods to screening tests for blood donation would be necessary to prevent malarial propagation through transfusion, particularly if the strain has a very long incubation period.
ACKNOWLEDGMENT
The authors thank the staff of Standard Diagnostics, Inc. (Yongin, Korea) and of Bioneer Co. (Daejeon, Korea) for their support and technical assistance.
