Different Clinical Outcomes of Entamoeba histolytica in Malaysia: Does Genetic Diversity Exist?
Article information
Abstract
The present study was conducted to investigate the clinical outcomes of Entamoeba histolytica infection in symptomatic and asymptomatic Orang Asli (aborigine) communities in Malaysia. Examination was performed on 500 stool samples obtained from Orang Asli communities in 3 different states using formalin-ether concentration, trichrome staining, and single-round PCR techniques. Out of 500 stool samples, single infection of E. histolytica, Entamoeba dispar, and Entamoeba moshkovskii was identified in 3.2%, 13.4%, and 1%, respectively. In addition, 10 samples had mixed infections with E. histolytica and E. dispar. Six samples containing E. dispar were also positive for E. moshkovskii, and only 2 samples had E. histolytica in association with E. dispar and E. moshkovskii. Seventeen E. histolytica-positive samples were from symptomatic subjects, whereas the remaining 11 samples came from asymptomatic subjects. These findings suggest a predominant distribution of pathogenic potential of E. histolytica strains in this community. Therefore, further studies on genotyping of E. histolytica is required, to find out association between E. histolytica genotype and the outcome of the infection.
Entamoeba histolytica is an enteric protozoan parasite that exists in either trophozoite or cyst form. The motile form (trophozoites) multiplies by binary fission and differentiates to the resistant form (cysts) that is responsible for transmission of the infection. Cysts are excreted in stools and may be ingested by a new host via contaminated water or food [1]. In the 1980s, the global prevalence of amebiasis was estimated to be approximately 10% of the world's population. Of these, approximately 90% were estimated to be asymptomatic carriers while only 10% will be developed to invasive amebiasis, leading to 110,000 deaths per year [2].
E. histolytica is capable of invading the intestinal mucosa and may spread to other extraintestinal organs, mainly the liver and rarely the kidneys, lungs, and brain. Thus, E. histolytica is unique among the intestinal amebae because it is able to invade tissue and clinical presentation may range from an asymptomatic infection to a disseminated fatal disease. Depending on the area of endemicity, the incubation period may vary from a few days to months [3]. Furthermore, even considering only E. histolytica infection, invasive amebiasis appears to be a relatively rare outcome of the infection. Therefore, the main objective of the present study was to investigate the clinical outcomes of E. histolytica infection in symptomatic and asymptomatic Orang Asli (aborigine) communities in Malaysia.
A total of 500 stool samples, comprising of 150 from Negeri Sembilan state, 139 from Perak state, and 211 from Pahang state in Peninsular Malaysia (Fig. 1) were collected over the period from June to December 2011. The participants were asked by a trained field assiatant to answer to a pre-tested questionnaire developed to elicit information on the demographic data, socioeconomic status, signs and symptoms, and medical treatment. After informed consent was obtained and questionnaire answered, all participants were then requested to provide a sufficiently large stool sample in a wide mouth screw-capped containers pre-labeled with their names and coded to enable both microscopic examination and molecular method to be performed. Approximately 5 g of each stool sample was kept into a 15-ml centrifuge tube containing 3 ml polyvinyl alcohol. The samples were subjected to modified trichrome staining [4], while another half of the samples were kept unfixed and stored at 4℃ upon arrival at the laboratory for further analysis by formalin-ether concentration [5]. Samples were considered microscopically positive if cysts and/or trophozoites of E. histolytica/dispar/moshkovskii were detected in at least 1 of the 2 techniques and negative if absent in all 2 techniques.
All stool samples were further characterized using molecular methods. Genomic DNA was extracted directly from all unfixed stool samples using QIAamp Stool DNA extraction kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The final DNA elution was made in 70 µl of elution buffer and stored at -20℃ until required for PCR amplification.
A single-round PCR assay and primer sets were used as described previously [6]. The sequence of the forward primer used (EntaF) was conserved in all 3 Entamoeba spp., whereas the specific reverse primers EhR, EdR, and EmR were specific for E. histolytica, E. dispar, and E. moshkovskii, respectively. The expected products were 166 bp (E. histolytica), 580 bp (E. moshkovskii), and 752 bp (E. dispar). DNA isolated from axenically grown E. histolytica HM-1:IMSS, E. dispar SAW 760, and E. moshkovskii Laredo was used as positive controls. All of these control DNA were a courtesy of Dr. Graham Clark (London School of Hygiene and Tropical Medicine, London, UK). Amplification of each species-specific DNA fragment started with an initial denaturation at 94℃ for 3 min, followed by 30 cycles of 94℃ for 1 min, 58℃ for 1 min and 72℃ for 1 min, with a final extension at 72℃ for 7 min. Its amplified products were analyzed by electrophoresis in 1.5% agarose gels and stained with GelRed (0.1 µl/ml, Biotium).
The PCR products were sequenced in both directions using the same primer sets as in the respective PCR assay using an ABI 3730XL sequencer (Bioneer Corporation, Seoul, Korea). Forward and reverse sequences were edited, manually aligned, and the consensus sequence was created for each sample using the BioEdit Sequence Alignment Search Tool (BLAST) to the National Centre for Biotechnology Information (NCBI) reference sequences (http://www.ncbi.nlm.nih.gov/BLAST). The E. histolytica (GenBank accession no. X56991) reference sequence was used in the analysis.
Prior to stool and data collections, the study protocol (reference no. UKM 1.5.3.5/244/FF-165-2011) was reviewed and approved by the Ethics Committee of Universiti Kebangsaan Malaysia Medical Centre (UKMMC) and permission for field works were obtained from the Ministry of Rural and Regional Development, Malaysia.
A total of 93 (18.6%) samples were microscopically positive for cysts and/or trophozoites of E. histolytica/dispar/moshkovskii, either singly or in combination with other protozoan parasites. Of the 93 microscopy-positive samples, single isolation of E. histolytica/dispar/moshkovskii was found in 19 (20.4%) samples, whereas the other 74 (79.6%) were mixed with 2 or more different protozoan species. Of the 500 Orang Asli who provided samples, 56.2% (281) were females and 43.8% (219) were males.
Of microscopy-positive stool samples containing E. histolytica/dispar/moshkovskii, PCR products were detected in 63 (67.7%) samples, whereas 30 (32.3%) were found to be negative (Table 1). Of the 30 PCR-negative samples, 13 were positive for cysts, and 17 contained both trophozoites and cysts of E. histolytica/dispar/moshkovskii. On the other hand, DNA products were detected in 10.6% (43/407) of microscopy-negative stool samples. Overall, PCR products were detected in 21.2% (106/500) of the tested samples (Fig. 2), whereas 78.8% (394/500) were found to be negative by a single-round PCR assay. Of the 106 PCR-positive samples, 3.2% were shown to contain single isolation of E. histolytica, 13.4% contained E. dispar, and only 1.0% contained E. moshkovskii. Mixed infections with E. histolytica and E. dispar were found in 2% of the samples. Six (1.2%) samples contained E. dispar and E. moshkovskii. PCR assay also detected 0.4% (2) of mixed infections by all the 3 species.
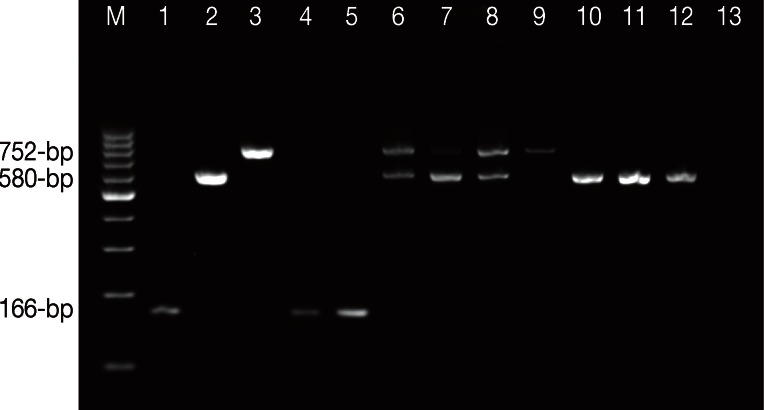
Agarose gel electrophoresis of Entamoeba spesies using single-round PCR. Lane M, 100-bp ladder DNA marker; Lane 1, E. histolytica HM-1:IMSS positive control; Lane 2, E. moshkovskii Laredo positive control; Lane 3, E. dispar SAW 760 positive control; Lanes 4 and 5, E. histolytica isolates; Lanes 6 to 8, mixed infections of E. dispar and E. moshkovskii isolates; Lane 9, E. dispar isolate; Lanes 10 to 12, E. moshkovskii isolates; Lane 13, negative control (DNAse free water, Fermentas).
Representative PCR products were sequenced in both directions from 16 samples of E. histolytica. Sequences of all 16 single isolations of E. histolytica showed high similarity (98-99%) to the E. histolytica sequences in GenBank (accession no. X56991). The 28 positive samples of E. histolytica were from 10 (35.7%) males and 18 (64.3%) females, aged between 2 and 61 years. Seventeen E. histolytica-positive samples were from symptomatic individuals, whereas the remaining 11 samples came from asymptomatic individuals. Out of 17, only 7 individuals had E. histolytica in association with pathogenic parasites such as Giardia intestinalis and soil-transmitted helminths (STHs) (Table 2).
E. histolytica is a common human pathogen that causes a spectrum of disease ranging from a commensal state in asymptomatic carriers to fulminant diarrhea or extraintestinal abscess formation. Indeed, less than 1 in 10 infections are now though to result in intestinal or extraintestinal symptoms in humans. The invasive strain of E. histolytica can cause ulceration of the intestinal epithelium and may penetrate the bowel wall to form extraintestinal abscess, especially in the liver. Several molecular types thought to correlate with the virulent phenotype have been partially characterized [7]. The pathogenic characteristic of several human E. histolytica isolates studied in different laboratories and tested in vivo and in vitro for their pathogenic capacity showed differences that can be associated with variations in the virulence potential between strains [8]. In addition, the advancements in molecular biology and genetics, now applied to amebic research, allow more precise search on intraspecies diversity markers associated with intestinal or extraintestinal invasive capacity of these protozoa in humans [9].
The present study demonstrated that 17 E. histolytica-positive samples were isolated from symptomatic subjects. The history of diarrhea, abdominal pain, vomiting, and other associated systemic symptoms such as fever, loss of appetite, and loss of weight were commonly seen in 61% individuals infected with E. histolytica. Likewise, a more recent study conducted in Turkey reported that almost all (13/14) of the children with E. histolytica infection had clinical symptoms, dysentery, cramping abdominal pain, diarrhea, flatulence, vomiting, and headache [10]. In the Netherlands, the researchers revealed that 3/4 of the E. histolytica carriers reported with abdominal complaints or diarrhea [11]. This finding is parallel with a study conducted in Sweden which demonstrated 10 patients that were diagnosed positive for E. histolytica showed different clinical manifestations ranging from diarrhea, abdominal pain, nausea, and constipation [12]. A study carried out in Egypt reported that the main symptoms among E. histolytica-dysenteric patients were colic and distention (64.3%), easy fatigue (57.1%) followed by tenesmus (50%), loss of weight (42.9%), and anorexia and vomiting (21.4%) [13], whereby the main presenting symptoms among non-dysenteric patients were colic and distention (66.7%) followed by fever (50%) and easy fatigue (16.7%). In Mexico, a study done by Redondo et al. [14] found a high correlation (98%) between clinical symptoms and E. histolytica infection and that the diagnosis of invasive amebae indicated that treatment should be done. The symptoms included dysentery, diarrhea, abdominal pain, and vomiting. Kebeda et al. [15] conducted a study among Ethiopian children showing that the most common complaints of E. histolytica infection were abdominal pain, tenesmus, mucoid bloody diarrhea, and distention, whereas fever, loss of weight, and constipation were less common.
However, there is a disparity between the present finding with previous studies which found a higher prevalence rate of E. histolytica asymptomatic infection in a rural Mexican community [16]. Likewise, a study conducted in the northern Philippines also reported that all of the E. histolytica-positive subjects were asymptomatic [17]. Furthermore, Haque et al. [18] reported that asymptomatic E. histolytica infection was common among preschool children in the urban slum of Dhaka, Bangladesh. This is consistent with several reports from endemic areas which showed that most E. histolytica infections are asymptomatic [19]. These findings were in agreement with the epidemiologic assertion before the characterization of E. histolytica and E. dispar species in the 1990s; 90% of E. histolytica-infected subjects are asymptomatic cyst passers [2].
In our study, diarrhea and other gastroenteritis symptoms were significantly associated with E. histolytica infection. The cause-effect relationship of E. histolytica with the clinical symptoms could not be determined in this present study due to the limitation of the design and no attempt to rule out other bacterial and viral infections. Besides these, it has long been known that not all E. histolytica infections lead to a clinical disease. The variables that are responsible for determining the different outcomes are still largely unknown. At present, we do not know whether some E. histolytica strains are intrinsically more virulent than others, but it has been reported that the outcome of E. histolytica infection may depend on several factors among which the genetic characteristics of the specific pathogen have been identified as an important factor. Few polymorphic genetic loci have been identified and targeted to aid in the study of the population structure of E. histolytica strains and their possible relationships with the parasite's virulence and disease outcome [20]. In order to investigate whether there is any link between the parasite and outcome of the infection, a reliable method for genotyping of the organism is required. Furthermore, in endemic areas of intestinal parasitic infections, mixed infections with STHs and other pathogenic protozoa that were also responsible for gastroenteritis symptoms were commonly observed. In this study, STHs, predominantly Trichuris trichiura, and Giardia intestinalis were detected in 42% (7/17) of symptomatic E. histolytica-positive subjects. Thus, these intestinal parasites may also account for the subjects' clinical symptoms.
In 30 samples positive for E. histolytica/dispar/moshkovskii by microscopy, we were unable to amplify DNA from any member of the E. histolytica/dispar/moshkovskii with the primers used and no inhibition of PCR was observed in control experiments. These results can potentially be explained by the presence of other Entamoeba species, which were detected by microscopy but not by PCR, or the presence of a low number of parasites in the sample, which fell below the detection limit of PCR. Another reason for this could be the fact that a majority of these samples (17/30) contained trophozoites that could have degenerated with time.
The discovery of low prevalence of E. histolytica infection in Orang Asli communities in this study indicates that this infection is not a public health problem in this community. However, identification of a high proportion of clinical symptoms among individuals positive for E. histolytica warrants further study to determine the causes. Therefore, immensely large scale, deliberate, and organized molecular genotyping studies are required to investigate whether there is any link between E. histolytica and the outcome of the infection in these communities.
ACKNOWLEDGMENTS
We sincerely thank Dr. Graham Clark (London School of Hygiene and Tropical Medicine, UK) for providing us with the lyophilized DNA of standard cultures of E. histolytica HM-1: IMSS, E. dispar SAW 760, and E. moshkovskii Laredo. We gratefully acknowledge the Ministry of Rural and Regional Development Malaysia for granting us permission to conduct this research.
We also thank all the participants from the Parit Gong village, Pasu village, Pian village, Bagan Balak village, Sungai Banun village, Desa Damai village, Sungai Raba village, and Pengkalan Permai village for their commitment and contribution in providing their stool samples.
This work was supported in part by the UKMMC Fundamental Research Grant (FF-165-2011) and Special Research University Grant (UKM-GUP-2011-316) from Universiti Kebangsaan Malaysia.