Sequence Analysis and Molecular Characterization of Wnt4 Gene in Metacestodes of Taenia solium
Article information
Abstract
Wnt proteins are a family of secreted glycoproteins that are evolutionarily conserved and considered to be involved in extensive developmental processes in metazoan organisms. The characterization of wnt genes may improve understanding the parasite's development. In the present study, a wnt4 gene encoding 491amino acids was amplified from cDNA of metacestodes of Taenia solium using reverse transcription PCR (RT-PCR). Bioinformatics tools were used for sequence analysis. The conserved domain of the wnt gene family was predicted. The expression profile of Wnt4 was investigated using real-time PCR. Wnt4 expression was found to be dramatically increased in scolex evaginated cysticerci when compared to invaginated cysticerci. In situ hybridization showed that wnt4 gene was distributed in the posterior end of the worm along the primary body axis in evaginated cysticerci. These findings indicated that wnt4 may take part in the process of cysticerci evagination and play a role in scolex/bladder development of cysticerci of T. solium.
INTRODUCTION
Cysticercus cellulosae is the causative agent of cysticercosis in both humans and pigs [1]. The disease is prevalent in many countries in Latin America, Africa, and Asia. It causes human neurocysticercosis and great economic loss in the pig-farming industry [2]. In endemic regions, transmission of this disease seems to be closely related to low standards of hygiene and environmental sanitation control [3,4]. The developmental process of cysticerci is such that when intermediate hosts such as pigs and humans incidentally ingest the eggs of Taenia solium, they hatch when exposed to the digestive enzyme in the duodenum. Then, oncospheres are released; the oncosphere penetrates the intestinal wall and is transported through the blood or lymphatics to tissues, where it develops into a cysticercus [1]. After the cysticercus established itself in tissues, the widening of the pore of the bladder wall allows the scolex and neck to evert. The mechanism underlying this event remains unclear thus far. Study of the molecular regulation mechanism of the developmental stages of cysticerci may facilitate the identification of potential drug targets and vaccine alternative antigens for efficient prevention and control of cysticercosis in humans and animals.
Wnt proteins are a family of secreted lipid-modified glycoproteins, typically 350-400 amino acids in length. They are evolutionarily conserved in metazoan organisms and are involved in a wide array of developmental processes and diseases [5]. They are characterized by the presence of 23-25 conserved cysteine residues [6]. They control diverse cellular behaviors, such as cell fate decisions, proliferation, and migration, and they are involved in many important embryological events, including axis specification, gastrulation, and limb, heart, or neural development via distinct signaling pathways [7]. A secreted wnt signal is typically transduced through 3 signaling pathways, the canonical wnt/β-catenin pathway, the wnt/planar cell polarity (PCP) pathway, and the wnt/Ca2+ pathway [8,9,10].
Wnt signaling in Schistosoma japonicum demonstrated that wnt4 regulated downstream gene expression by the canonical pathway [11]. A similar signaling pathway was predicted in the genome of T. solium [12]. Characterization of wnt4 of cysticercus may provide valuable insight into the complicated developmental mechanism of this parasite.
MATERIALS AND METHODS
Parasite
A 3-month-old pig was drenched with solution containing eggs, which were derived from a T. solium-infected adult human patient from Jilin Province, China. Ninety days later, cysticerci were collected from the muscles of the pig after killing and dissection. Animal experiments were performed on this pig only after formal ethical approval, which was in accordance with the facility's institutional and governmental guidelines. The larvae obtained in this way were washed 3 times with normal RNase-free saline water. Some of the larvae were cultivated for the scolex evagination by incubating in 5 ml of pig bile buffer, which was 1:3 diluted in RPMI 1640 medium (Gibco, Grand Island, New York, USA) for 2 hr at 37℃. Both the evaginated and invaginated cysticerci were stored at RNAlater RNA Stabilization Reagent (Qiagen, Venio, Netherlands) in -70℃ until use.
Preparation of total RNA from cysticerci
The frozen cysticerci were ground into powder in liquid nitrogen. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions. Any residual DNA that remained in the RNA solution was excluded by digestion using RNase-free DNaseI (Promega, Madison, Wisconsin, USA). The integrity of the RNA was verified by 1% non-denatured agarose gel electrophoresis, and recovered RNA was quantified and qualified using a Thermo Scientific NanoDrop 2000 spectrophotometer (Thermo Scientific, Hudson, New Hampshire, USA).
PCR amplification
Reverse transcription PCR (RT-PCR) was conducted using freshly prepared RNA and Reverse Transcription System (Promega) according to the manufacturer's instructions. The synthesized cDNA was used for the PCR amplification with specific primers (50 µl) (Reverse Transcription System, Promega), F: 5'-TAATGAACTCCGCACCATGCT-3'; R: 5'-TTTTTTCACATGTCTCGCATCG-3', which were designed based on the wnt4 gene reference sequence derived from the T. solium genome database (www.192.168.51.199). The cycling conditions were as follows: denaturation at 94℃ for 5 min, followed by 35 cycles of denaturation at 94℃ for 30 sec, annealing at 52℃ for 40 sec and extension at 72℃ for 90 sec. This was followed by a final extension of 72℃ for 10 min. The PCR product was analyzed on 1% agarose gel. The 5'-RACE (rapid amplification of cDNA ends) and 3'-RACE were performed using the SMART™ RACE cDNA Amplification Kit (Clontech, Tokyo, Japan) according to the manuals. The gene specific primer for the 5'-RACE was 5'-GTG-CTGACTGTTGGAGGCATTGGAT-3'. The specific primer for the 3'-RACE was 5'-GCGACTACCGCAGAGAGGTGTTCAT-3'. All the primers were synthesized by Sangon Biotech Ltd. (Shanghai, China).
The RT-PCR and RACE products were ligated to the pMD18-T vector (TaKaRa, Dalian, China), and then transformed into Escherichia coli DH5α competent cells. The positive clones were selected for sequencing (Sangon, Shanghai, China).
Bioinformatic analysis
The wnt4 fragments from PCR, 5'-RACE, and 3'-RACE were assembled into a complete cDNA sequence using DNASTAR software (Madison, Wisconsin, USA). The homologous genes were searched and compared using BLASTn/p in NCBI (http://www.ncbi.nlm.nih.gov). The gene structure was analyzed using GSDS (http://gsds.cbi.pku.edu.cn/) on the basis of the cDNA sequence and the DNA sequence from the T. solium genome database (www.192.168.51.199). Signal peptides were predicted using Signal P3.0 (http://www.cbs.dtu.dk/services/SignalP/). The related wnt4 amino acid sequences were obtained from GenBank and Sanger databases for alignments. Alignments was performed using ClustalW (http://www.ebi.ac.uk/clustalw/). The post-translational modification of wnt4 protein was analyzed using Proscan (http://npsapbil.ibcp.fr/).
qRT-PCR
The expression profiles of wnt4 were probed using qRT-PCR in scolex evaginated and invaginated cysticerci. The template cDNAs were synthesized using the 2-step MMLV Platinum® SYBR® Green qPCR SuperMix-UDG w/ROX (Invitrogen) according to the user manuals. The primers for wnt4 were F: 5'-AGGAAGCCAGGTAAAGTGCTC-3' and R: 5'-CAGCCAGACCACTCAAAGTTG-3'. The glyceraldehydes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene served as a control. It was amplified using F: 5'-GGCATCAATACCACCATC-3' and R: 5'-GGAAGGACCATCAACAAC-3'.
The qRT-PCR was performed using a MX3005P (Agilent, Palo Alto, California, USA) according to the manufacturer's instructions. Each sample was tested in triplicate. The cycling conditions comprised preincubation at 50℃ for 2 min and denaturation at 95℃ for 2 min, followed by 45 cycles of 95℃ for 15 sec, 55℃ for 30 sec, 72℃ for 30 sec. A serial dilution of GAPDH cDNA served for establishment of the standard curves.
RNA in situ hybridization
Synthesis of cRNA probe
A wnt4 fragment with 248 bp in size was amplified using primers F: 5'-AGGAAGCCAGGTAAAG TGCTC-3' and R: 5'-CAGCCAGACCACTCAAAGTTG-3'. The positive products were cloned in the pGEM-Teasy vector (Promega) and then transformed into E. coli JM109 cells for preparation of the plasmid. The resultant plasmid was linearized using NdeI and SacII endonuclease digestion, and then purified. RNA labeling with digoxigenin-UTP by in vitro transcription with SP6 and T7 RNA polymerase was performed using SP6 DIG RNA Labeling Kit (SP6/T7) (Roche, Mannheim, Switzerland) according to the manufacturer's instructions. The riboprobes were then purified with ethanol precipitation.
In situ hybridization
The fresh scolex evaginated and invaginated cysticerci were washed 3 times with RNase-free saline water, and then fixed with 4% formaldehyde (pH 7.2) in RNase free PBS at 4℃ overnight. The cysticerci were treated with gradient ethanol dehydration, which included subsequent immersing shortly in each of 35%, 50%, 70%, and 90% and twice in 100% ethanol; in 50% ethanol and 50 % xylene mixed solution; in 100% xylene, in 50% xylene and 50% paraffin mixed solution; and 3 times in paraffin. Then, the dehydrated larvae were embedded in paraffin wax. Slices were prepared from the paraffin wax using a Superfrost plus microscope slides (Fisher Scientific, Silver Spring, Maryland, USA). The slices were flatly placed onto glass slides and then incubated at 42℃ overnight. The slides were dewaxed by 2 rounds of washes with xylene and rehydrated twice in 100% ethanol and then once each in 90%, 70%, 50%, and 35% ethanol. The slides were rinsed twice in DEPC water and then treated with proteinase K (1 µg/ml) at 37℃ for 5 min. The proteinase K was blocked for 2 min with glycine.
The slides were then treated with the following steps: washed with PBS; fixed with 4% paraformaldehyde (PFA) for 10 min; washed with PBS; incubated for 20 min with 0.2 M HCl; and washed twice with PBS. The slides were then incubated with hybridization buffer (50% formamide, 1×SSC, 0.5 mg/ml yeast tRNA, 10% dextran sulfate, 10 mM DTT) for 2 hr at 55℃ for pre-hybridization, followed by overnight incubation with 5 ng/µl digoxigenin-labeled probe. Then, 3 high-stringency washes were performed with 0.2× SSC at 62℃. The excess probe was digested with 20 µg/ml RNAse at 37℃ for 30 min. Slides were incubated with horseradish alkaline phosphatase (AP)-conjugated anti-digoxigenin, and then using NBT (nitrobluetetrazolium)/BCIP (5-bromo-4-chloro-indolyl phosphate) for signal development according to DIG nucleic acid detection kit (Roche).
RESULTS
Sequence analysis of T. solium wnt4
The assembled full wnt4 was 1,473 bp in length with an open reading frame encoding a 491 amino acid protein (GenBank accession no. KF700085). It had a 311 bp 3' untranslated region (UTR) and a 66 bp 5' UTR. Signal P3.0 server prediction (http://www.cbs.dtu.dk/services/SignalP/) revealed a signal peptide of 23 amino acids at the N terminus. There were 23 cysteine residues and 2 presumed N-glycosylation sites. Analysis of the wnt4 post-translational modification in multiple species showed there to be 3 N-myristoylation sites in free-living Schmidtea mediterranea, and more than 10 N-myristoylation sites in Schistosoma japonicum, S. mansoni, Echinococcus multilocularis, and T. solium. The same motif C-[KR]-C-H-G-[LIVMT]-S-G-x-C was found in the platyhelminthes and the intermediate host (Homo sapiens and Sus scrofa) of T. solium (Table 1).
Differential expression of wnt4 in scolex evaginated and invaginated cysticerci
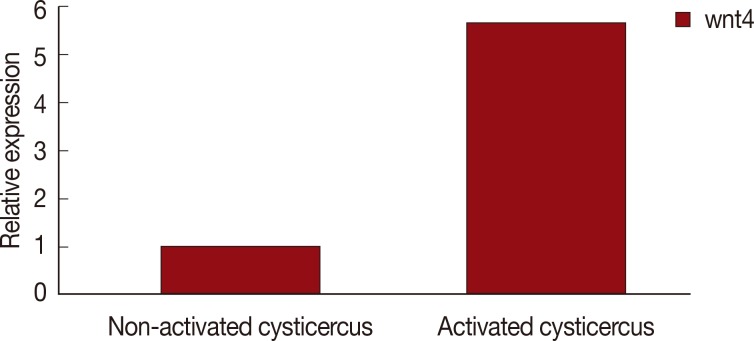
In the qRT-PCR, the efficiency of the primers specifically for wnt4 and GAPDH were found to be within an acceptable range of 97.2-104.4%. The standard curves of wnt4 and reference genes showed a linear regression coefficient of determination (R2) between 0.991 and 0.999. The expression level of wnt4 gene in different stages of scolex evaginated and invaginated cysticerci were determined using qRT-PCR and the 2-ΔΔCT method. As shown in Fig. 1, the wnt4 gene expression was significantly upregulated (P<0.01, 5.7-fold) in the scolex evaginated cysticerci.
Localization of wnt4 in scolex evaginated cysticercus
The wnt4 gene was deeply expressed in the bladder wall of the scolex evaginated cysticercus as shown in Fig. 2A, but no reaction was observed in the scolex invaginated cysticercus. Neither the scolex evaginated nor invaginated cysticerci were stained with the sense probe.
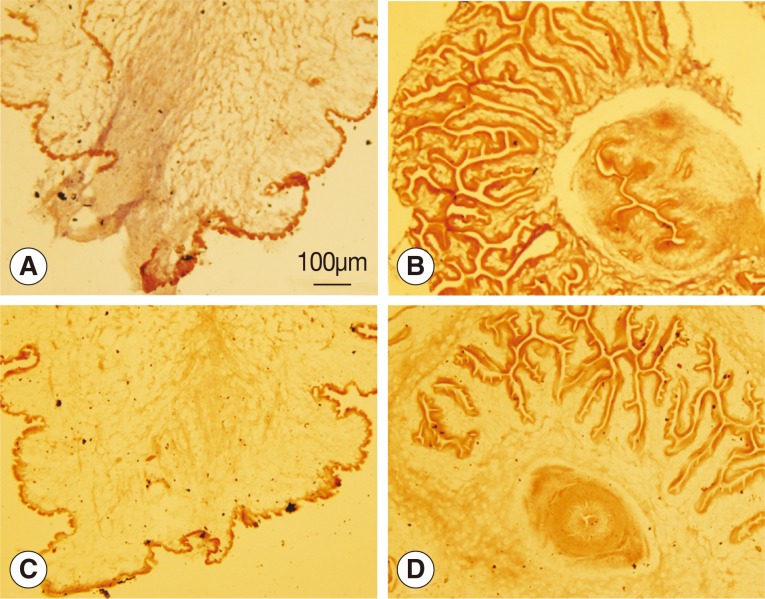
Analysis of wnt4 expression by in situ hybridization. (A, B) Antisense probes were used. (C, D) Sense probes were used. (A, C) Scolex evaginated cysticercus. (B, D) Scolex invaginated cysticercus. The expression of wnt4 was deeply stained from the posterior end along the primary body axis of cysticercus as shown in A.
DISCUSSION
Wnts are secreted glycoproteins that function as extracellular signals across a wide range of metazoans, including cnidarians, nematodes, insects, and vertebrates [13]. Phylogenetic analyses group them into 13 subfamilies: wnt-1 through wnt-11, wnt-16, and wnt-A [14]. Flatworms have a highly reduced and dispersed complement that includes orthologs of only 5 subfamilies (wnt1, wnt2, wnt4, wnt5, and wnt11). This is less than in planarians (9 species) [15]. Wnt1, wnt2, wnt4, wnt5, wnt11a, and wnt11b were found during T. solium genome analyses (data not shown). Sequence alignments showed that the parasitic flatworms (Echinococcus granulosus, E. multilocularis, Hymenolepis microstoma, S. mansoni, and T. solium) possess a single ortholog of wnt4.
Post-translational modification of wnt proteins plays critical roles in many aspects of wnt regulation, including ligand secretion, extracellular distribution and receptor activation [16]. Analysis of wnt4 post-translational modifications in parasites and planarians revealed a tyrosine kinase phosphorylation site and a cAMP- and cGMP-dependent protein kinase phosphorylation site, neither of which are present in humans or pigs (Table 1). Phosphorylation is essential for intracellular and extracellular signals that are transmitted throughout cells [17,18]. Studies have shown that phosphorylation events play an important role in growth, development, immunity, and survival in parasitic flatworms with assistance of receptor tyrosine kinases (RTKs) [18,19,20,21,22]. More than 10 N-myristoylation sites were found in wnt4 of T. solium, other parasitic flatworms, H. sapiens, and S. scrofa; but only 3 sites were found in planarians. Myristoylation is functional in the regulation of mitochondrial integrity and generation of pro-death or pro-survival signals [23]. Both phosphorylation and myristoylation are keys to the cell's ability to make life and death decisions. The post-translational modifications of host-parasite relationships raises interesting questions regarding the impact of parasitism on the co-evolution of ligand and receptor molecules in helminth parasites [18].
The qPCR showed that wnt4 to be upregulated in the scolex evaginated cysticerci, indicating that wnt4 is likely to be related to larval growth. This is because it has shown that wnt signaling is involved in a diverse range of cellular interactions throughout development, including regeneration, segmentation, and axial patterning [24,25,26,27,28]. The wnt4 pathway has shown that wnt4 of S. japonicum mediates signal transduction by the canonical wnt/β-catenin pathway, suggesting that wnt4 takes part in inducing axis duplication and/or displaying transforming activity [11]. It is here speculated that wnt4 of T. solium is involved in the canonical pathway because of its high level of expression in the scolex evaginated larvae.
The wnt canonical pathway signaling is responsible for the regulating of head versus tail development during planarian regeneration [29,30,31,32]. Wnt4 was here found to be expressed in the posterior ends of the worms along the primary body axis in the scolex evaginated cysticercus, indicating that wnt4 of T. solium is involved in the canonical pathway, and it is likely functional in the evolution of segmentation in platyhelminthes.
In conclusion, this is the first study of identification and characterization of wnt4 gene of metacestodes of T. solium. Sequence analysis revealed specific post-translational modification features. A high level of wnt4 gene expression in the scolex evaginated cysticercus indicated that wnt4 likely plays an important role in the process of cysticercus evagination. Localization demonstrated that wnt4 was distributed mainly from the posterior end along with the primary body axis in the scolex evaginated cysticercus. This supports the assumption that wnt4 is functional in the evolution of segmentation in platyhelminthes.
ACKNOWLEDGMENTS
We are grateful to Dr. Yadong Zheng for his kind advice. We also thank Dr. Xiaolin Sun for his technical assistance in slices preparation. This work was supported by the National Natural Science Foundation of China (Grant no. 31201898), Science Fund for creative research groups of Gansu Province (Grant no. 1210RJIA006), and Open Funds of the state key laboratory of veterinary etiological biology.
Notes
We declare that we have no conflict of interest related to this study.