Molecular Analysis of Anisakis Type I Larvae in Marine Fish from Three Different Sea Areas in Korea
Article information
Abstract
Anisakiasis, a human infection of Anisakis L3 larvae, is one of the common foodborne parasitic diseases in Korea. Studies on the identification of anisakid larvae have been performed in the country, but most of them have been focused on morphological identification of the larvae. In this study, we analyzed the molecular characteristics of 174 Anisakis type I larvae collected from 10 species of fish caught in 3 different sea areas in Korea. PCR-RFLP and sequence analyses of rDNA ITS and mtDNA cox1 revealed that the larvae showed interesting distribution patterns depending on fish species and geographical locations. Anisakis pegreffii was predominant in fish from the Yellow Sea and the South Sea. Meanwhile, both A. pegreffii and A. simplex sensu stricto (A. simplex s.str.) larvae were identified in fish from the East Sea, depending on fish species infected. These results suggested that A. pegreffii was primarily distributed in a diverse species of fish in 3 sea areas around Korea, but A. simplex s.str. was dominantly identified in Oncorhynchus spp. in the East Sea.
INTRODUCTION
Anisakiasis (or Anisakidosis) is a fishborne parasitic infection, which is caused by an accidental infection with larval nematodes belonging to the family Anisakidae when consuming raw, undercooked, or improperly processed marine fish and cephalopods, resulting in sudden gastrointestinal disorders, nausea, vomiting, diarrhea, or allergic reactions [1]. Human anisakiasis has been increasing worldwide in accordance with the increase of consumption of raw or undercooked marine fish [2,3]. Most of the human cases have been reported in East Asia, including Japan and Korea, but there have also been increasing reports from other geographical areas or countries, including coastal areas of Europe, New Zealand, Canada, Brazil, Chile, and Egypt [2,3,4]. Anisakis simplex (Anisakis type I), A. physeteris (Anisakis type II), and Pseudoterranova decipiens (Terranova type A) larvae have been identified as the major causative agents of human anisakiasis [1,4].
Several studies on the infection status of anisakid larvae in marine fish and cephalopods have been performed in Korea. Various types of anisakid larvae in the genus Anisakis, Contracaecum, Raphidascaris, and Hysterothylacium were identified in a variety of marine fish and cephalopods caught in the coastal areas of Korea [5,6,7,8]. Based on morphological characteristics, A. simplex (Anisakis type I) is the most dominant species among anisakid larvae found in fish paratenic hosts and human infection cases, but species identification at the molecular level is necessary to clarify the species identity between the closely related ones, such as A. simplex sensu stricto (s.str.), A. pegreffii, A. typica, A. physeteris, and so on [4].
In this study, we analyzed the molecular characteristics of Anisakis type I larvae collected from 10 fish species caught in 3 different locations of Korea to obtain information on distribution and paratenic hosts of Anisakis species in the sea areas of the country.
MATERIALS AND METHODS
Parasites
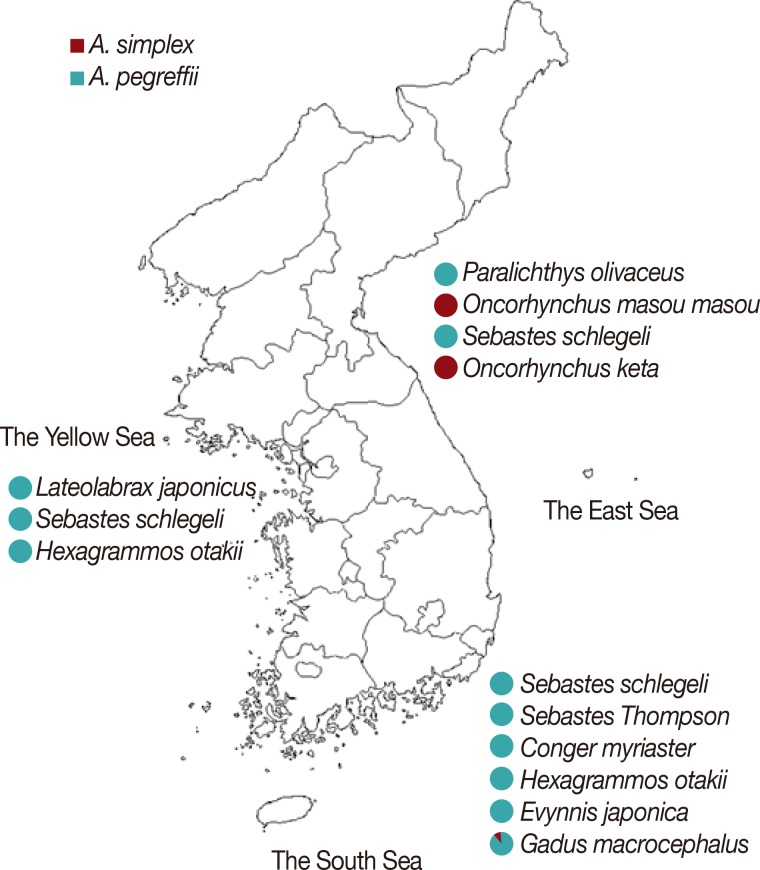
Anisakis type I larvae (n=174) used in this study were collected from 10 species of marine fish purchased from 3 geographically different local fish markets in Goseong-gun, Gangwon-do (the East Sea), Sacheon City, Gyeongsangnam-do (the South Sea), or Incheon Metropolitan City (the Yellow Sea) in 2008 [7]. The parasites were morphologically confirmed as Anisakis type I larvae under stereomicroscopic examination [7]. The number of larvae isolated from each species of fish is summarized in Fig. 1.

Anisakis type I larvae used in this study. A total of 174 larvae randomly selected from morphologically confirmed Anisakis type I larvae, isolated from 10 species of fish caught in 3 different sea areas in Korea, were used. The number of larvae isolated from each species of fish and analyzed in this study is presented. ●, fish purchased sites.
Amplifications of rDNA ITS and mtDNA cox1 regions
Genomic DNA was extracted from individual larva using a Genomic DNA Extraction Kit (Real Biotech Corporation, Banqiao City, Taiwan) according to manufacturer's instruction. Amplifications of rDNA ITS (ITS1, 5.8 subunit rRNA gene, and ITS2) and mtDNA cox1 were performed by PCR. The primers used for the amplification of ITS were 5'-GTCGAATTCGTAGGTGAACCTGCG-3' and 5'-GCCGGATCCGAATCCTGGTTAGTT-3' [9]. For amplification of cox1, 5'-TTTTTTGGGCATCCTGAGGTTTAT-3' and 5'-TAAAGAAAGAACATAATGAAAATG-3' were used [10]. The amplification reactions for ITS and cox1 were performed using the following thermal cycling conditions: 94℃ for 5 min, 30 cycles at 94℃ for 1 min, 52℃ for 1 min, and 72℃ for 1 min, followed by a final extension at 72℃ for 10 min. In order to minimize the likelihood of possible nucleotide misincorporations in the amplification processes, Ex Taq DNA polymerase (Takara, Otsu, Japan), which has a proof-reading function, was used in all the PCR reactions.
RFLP analysis
Each PCR product was digested with 10 units of each restriction endonuclease, Hinf I (Takara) or Hha I (Takara), respectively, in a volume of 20 µl at 37℃ for 3 hr [11]. The reaction mixtures were separated on 2.5% agarose gels containing Red Safe™ nucleic acid staining solution (Intron Biotechnology, Sungnam, Korea) and the restriction fragments were visualized under a UV illuminator.
Sequencing and phylogenetic analysis
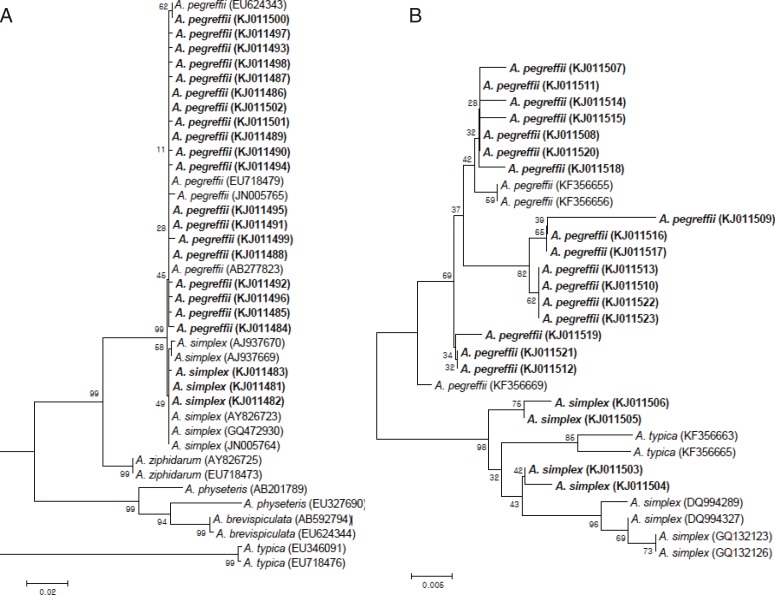
The PCR product was analysed on a 1.2% agarose gel, purified from the gel, and then ligated into the T&A cloning vector (Real Biotech Corporation). Each ligation mixture was transformed into Escherichia coli DH5α competent cells, and positive clones with the appropriate insert were selected by colony PCR. The nucleotide sequences of the cloned inserts were analysed by automatic DNA sequencing. Nucleotide and deduced amino acid sequences were analyzed using EditSeq and Clustal in the Megalign program, a multiple alignment program in the DNASTAR package (DNASTAR, Madison, Wisconsin, USA). The nucleotide sequences for ITS and cox1 were registered in the GenBank database (accession nos. KJ011481-KJ011523). The phylogenetic trees for ITS and cox1 were constructed using the neighbor-joining method in MEGA 4 [12]. Bootstrap proportions were used to assess the robustness of the tree with 1,000 bootstrap replications.
RESULTS
PCR-RFLP patterns of rDNA ITS
The 174 Anisakis type I larvae collected from 10 fish species caught in 3 different sea areas in Korea were analyzed by PCR-RFLP on the basis of the restriction fragment patterns. Amplification of the ITS region produced an amplicon of approximate size of 1,000 bp in all analyzed samples. Digestion of the amplified ITS with Hinf I resulted in 2 different fragment patterns corresponding to 2 sibling species of A. simplex complex, A. simplex s.str. and A. pegreffii (Fig. 2). The banding patterns observed in this study were well corresponded to the known patterns of A. pegreffii and A. simplex s.str. reported previously [6,11]. Digestion with Hinf I produced 2 strong bands and a weak band (620, 270, 80 bp, respectively) for A. simplex s.str., whereas A. pegreffii showed 3 fragments of 370, 300, and 250 bp (Fig. 2A). Meanwhile, digestion with Hha I yielded the same digestion pattern of 2 fragments of approximately 550 and 430 bp for all the samples tested (Fig. 2B).
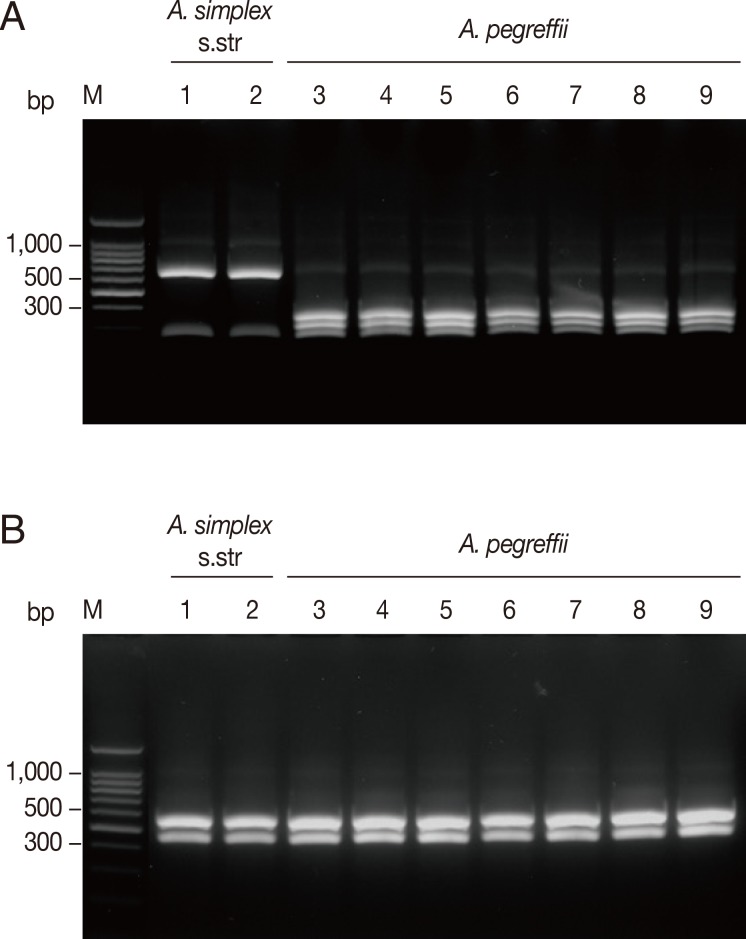
PCR-RFLP analysis of rDNA ITS for molecular identification of anisakid larvae. Each PCR product was digested with Hinf I or Hha I and analyzed on agarose gel. (A) Hinf I digestion. (B) Hha I digestion. Lanes 1-2, A. simplex s. str. (each from G. macrocephalus and O. keta, respectively); Lanes 3-9, A. pegreffii (each from L. japonicus, S. thompson, C. myriaster, H. otakii, E. japonica, G. macrocephalus, and P. olivaceus, respectively); Lane M, 100 bp ladder.
PCR-RFLP patterns of mtDNA cox1
Amplification of the mtDNA cox1 produced an amplicon of about 490 bp in all analyzed samples. The RFLP patterns with Hinf I digestion produced different fragment patterns depending on the species of anisakids (Fig. 3). A. simplex s.str. showed 2 fragments of approximately 270 and 210 bp, while A. pegreffii showed 2 strong fragments of approximately 270 and 150 bp and a weak fragment of approximately 80 bp. The banding patterns observed in this study were also well corresponded to the patterns of A. pegreffii and A. simplex s.str. reported previously [11].

PCR-RFLP analysis of mtDNA cox1 for molecular identification of anisakid larvae. Each PCR product was digested with Hinf I and analyzed on agarose gel. Lanes 1-3, A. pegreffii (each from S. schlegeli, C. myriaster, and H. otakii, respectively); Lanes 4-6, A. simplex s. str. (each from G. macrocephalus, O. masou masou, and O. keta, respectively); Lane M, 100 bp ladder.
Sequence and phylogenetic relationships
We also performed sequencing analysis of ITS and cox1 of randomly selected larvae from different species of fish to ensure the performance of PCR-RFLP. Based on the presence of Hinf I and Hha I restriction sites in the sequences, all the sequences obtained were well corresponded to the results by PCR-RFLP analysis (data not shown). The ITS gene sequences of A. pegreffii and A. simplex s.str. isolated from fish analyzed in this study showed 98.6-100% and 98.6-99.8% similarities with those of previous reported sequences, respectively. The cox1 analyzed here also showed high sequence similarities of 96.3-99.7% and 96.7-98.7% with those of previously reported A. pegreffii and A. simplex s.str., respectively. The phylogenetic analyses of both ITS and cox1 revealed that the taxonomic positions for the Anisakis species in the phylogenetic tree corresponded well to the identification results by PCR-RFLP analysis (Fig. 4).
Distribution of A. simplex complex in each species of fish
PCR-RFLP and sequence analyses of ITS and cox1 revealed that all 48 larvae isolated from the 3 species of fish caught in the Yellow Sea were A. pegreffii (Fig. 5). Most of the larvae collected from 6 species of fish caught in the South Sea were also A. pegreffii except only 1 larva isolated from Gadus macrocephalus, which was identified as A. simplex s.str. The most interesting finding was seen in the larvae collected from fish caught in the East Sea. All the larvae infected in Paralichthys olivaceus and Sebastes schlegeli were identified as A. pegreffii. On the other hand, all the larvae collected from Oncorhynchus masou masou and Oncorhynchus keta were A. simplex s.str. No mixed infection of A. simplex s.str. and A. pegreffii was identified in all fish used in this study.
DISCUSSION
Based on their morphological features, larvae of the genus Anisakis can be identified as Anisakis type I or type II. They differ in the length of their ventriculus and in the presence of mucron at the posterior end [13]. These morphological analyses have been widely used for classification of anisakid nematodes [13,14], but morphological characteristics are not always consistent and are insufficient for species identification [15]. With regard to the genus Anisakis, it has been reported that Anisakis type I larvae (morphological type) comprise several species with different genetic characters, including A. simplex s.str., Anisakis pegreffii, and A. simplex C [16]. Thus, molecular approaches have been introduced for accurate classification and the study of the systematic evolution of anisakid nematodes [6,9,11,15,17,18].
Here, we identified 2 sibling species of A. simplex complex, A. simplex s.str. and A. pegreffii, in 10 fish species caught in 3 different sea areas in Korea and analyzed their distributions in the paratenic hosts. Both RFLP and sequencing analyses of ITS and cox1 revealed that A. pegreffii was predominant in 7 fish species from the Yellow Sea and the South Sea. This species has been reported as the dominant species identified in several fish and squid from the South Sea and the Yellow Sea of Korea [6,8] as well as the East China Sea and sea areas of southern Japan [11,19,20,21]. Only 1 A. simplex s.str. was identified among 10 Anisakis type I larvae, and it was collected from Gadus macrocephalus caught in the South Sea. The other 127 larvae collected in 7 fish species caught in the Yellow Sea and the South Sea were A. pegreffii.
Meanwhile, in the cases of larvae collected from fish caught in the East Sea, an interesting finding was observed. Among the 4 fish species caught in this area, 2 of them, Paralichthys olivaceus and Sebastes schlegeli, were infected only with A. pegreffii. Meanwhile, all the larvae collected from the other 2 species of fish, O. masou masou and O. keta, were determined to be A. simplex s.str. Our results are consistent with previous reports, which indicate predominant infection of A. simplex s.str. in Oncorhynchus spp. [6,22].
Occurrence of hybrid genotypes, which were probably produced by natural interspecific hybridization between A. simplex s.str. and A. pegreffii, was recently reported in several studies [8,11,23,24]. However, no clear hybrid genotype of larvae was identified in this study. Considering the low levels of hybrid type of larvae have been identified in the previous studies [8,11,23,24], limited numbers of anisakid larvae analyzed in this study can be a possible factor. Further extensive molecular characterization of larger number of anisakid larvae in various fish hosts, with distinct geographical and seasonal information, will be necessary to clarify the presence and infection status of larvae with hybrid genotypes.
A. simplex s.str. has been recognized as the major causative agent of human anisakiasis. Although A. pegreffii comprise the abundance of larvae identified in fish caught offshore in Japan, the main species of larvae associated with human anisakiasis have been identified as A. simplex s.str. [25]. Moreover, A. pegreffii is likely to be irrefutably pathogenic to humans, since only a few clinical cases caused by A. pegreffii infections have been reported worldwide [26,27]. Several studies have presented interesting results on the pathogenic differences between the Anisakis species. A. simplex s.str. larvae can survive for a longer time in acidic artificial gastric juice and has higher agar penetration activity than A. pegreffii larvae [16,28]. In addition, A. simplex s.str. larvae are likely to possess stronger tissue-invasive traits relevant not only to fish musculature but also to the human body [22]. These suggest that pathogenetic differences between Anisakis species exist, even though these differences require further elucidation. Many clinical cases of human anisakiasis have also been reported in Korea [29,30,31], but molecular classification of the infected larvae was not performed. Therefore, further studies on the identification of anisakid species in human anisakiasis are warranted to clarify the relevance of anisakid species involved in human anisakiasis.
In conclusion, our results collectively suggest that 2 sibling species of anisakid nematodes are distributed in different species of fish in Korean sea waters. A. pegreffii is the most prevalent species detected in the fishes caught offshore in Korea, especially those in the South Sea and the Yellow Sea, but all the larvae collected from O. masou masou and O. keta in the East Sea were A. simplex s. str.. These data provide new information on the distribution and paratenic hosts of the anisakid larvae in Korean coastal areas as well as fundamental epidemiological and biological information on anisakid species. Our results also emphasize the potential hazards of anisakid larvae for human health as a consequence of consuming raw or undercooked fish. Considering the high occurrence and infection burden of anisakid larvae in the fish caught offshore in Korea [5,7], further comprehensive studies to determine the infection status of Anisakis larvae in humans as well as in other relevant commercial fish species is necessary.
Notes
We have no conflict of interest related to this study.