Genetic Diversity of Echinococcus granulosus in Center of Iran
Article information
Abstract
Hydatid cyst caused by Echinococcus granulosus is one of the most important parasitic diseases around the world and many countries in Asia, including Iran, are involved with this infection. This disease can cause high mortality in humans as well as economic losses in livestock. To date, several molecular methods have been used to determine the genetic diversity of E. granulosus. So far, identification of E. granulosus using real-time PCR fluorescence-based quantitative assays has not been studied worldwide, also in Iran. Therefore, the aim of this study was to investigate the genetic diversity of E. granulosus from center of Iran using real-time PCR method. A total of 71 hydatid cysts were collected from infected sheep, goat, and cattle slaughtered in Isfahan, Iran during 2013. DNA was extracted from protoscolices and/or germinal layers from each individual cyst and used as template to amplify the mitochondrial cytochrome c oxidase subunit 1 gene (cox1) (420 bp). Five cattle isolates out of 71 isolates were sterile and excluded from further investigation. Overall, of 66 isolates, partial sequences of the cox1 gene of E. granulosus indicated the presence of genotypes G1 in 49 isolates (74.2%), G3 in 15 isolates (22.7%), and G6 in 2 isolates (3.0%) in infected intermediate hosts. Sixteen sequences of G1 genotype had microgenetic variants, and they were compared to the original sequence of cox1. However, isolates identified as G3 and G6 genotypes were completely consistent with original sequences. G1 genotype in livestock was the dominant genotype in Isfahan region, Iran.
INTRODUCTION
Hydatid cyst, caused by Echinococcus granulosus is one of the most important parasitic diseases worldwide and distribute in many Asia countries, including Iran. This disease can cause high mortality in humans as well as economic losses in livestock [1,2,3]. Nowadays, genetic studies on a partial sequence of mitochondrial cytochrome c oxidase subunit 1 (cox1) and nuclear genetic markers suggest that E. granulosus has 10 distinct genotypes termed G1-G10 [4,5,6,7,8,9]. These different genotypes include G1 and G2 as sheep strains, G3 and G5 as bovine strains, G4 and G6 as horse and camel strains, G7 as a pig strain, G8 and G10 as cervid strains [10]. Recent taxonomic studies have suggested E. granulosus in 4 distinct groups, including E. granulosus sensu stricto (G1-G3 genotypes), Echinococcus equinus (G4), Echinococcus ortleppi (G5), and Echinococcus canadensis (G6-G10) [11]. The identification of E. granulosus strains has been carried out in different laboratories using various analytical methods (morphology, physiology, biochemistry, and molecular genetics), and all of them were proved to be useful, particularly when used together [12,13,14,15,16]. The possible susceptibility factors for genetic variation that occur in E. granulosus may influence many different phenotypic features such as the life cycle patterns, development rate, host specificity, geographical distribution, transmission dynamics, infectivity, antigenicity, control of diseases, and chemotherapeutic agents.
PCR-based methods have been extensively used to characterize the different strains of E. granulosus [12]. On the other hand, various techniques such as PCR-RFLP, nested PCR, and real-time PCR were used to determine genetic variations within E. granulosus. However, fluorescence-based quantitative PCR assays has not fully developed yet [17,18]. Real-time PCR is a combination of PCR chemistry with fluorescent probe for detection of amplicon which shows results with curve [19]. Specifically, EVA Green™-based real-time PCR, in association with melting curve analysis, can be useful for detection of nucleotide variation such as single nucleotide polymorphism (SNP). Different PCR products often have a different melting temperature (Tm), and this Tm is related to the percentage of guanine-cytosine (GC), length, and nucleotide composition [20]. In Iran, especially in Isfahan, sheep is the most common and important intermediate host for E. granulosus. According to previous studies, the prevalence of infection and cyst fertility rates were higher in sheep than in other slaughtered animals [21,22,23]. None of the previous studies in Iran was carried out using real-time PCR method for detection of E. granulosus. Therefore, the aim of this study was to investigate the genetic diversity of E. granulosus from center of Iran using real-time PCR method.
MATERIALS AND METHODS
Sample collection
Seventy-one hydatid cysts were collected from infected slaughtered animals in different regions of Isfahan, including Fasaran, Khomeinishahr, and Najafabad from January 2013 until March 2013. Separation of hydatid cysts was performed under sterile condition, and then protoscolices and/or the germinal layer were collected from an individual hydatid cyst. Protoscolices and the germinal layer were preserved in 70% (v/v) ethanol and stored at -20℃ until DNA extraction.
DNA extraction
Before DNA extraction, ethanol was removed by 3 times washing with sterile distilled water. Total genomic DNA was extracted using a genomic DNA extraction kit (Bioneer, Daejeon, Korea) according to the manufacturer's instructions with some modifications. The concentration of extracted DNA was evaluated by Nano Drop, and then the samples were kept at -20℃ for further analysis.
PCR and DNA sequencing
Genomic DNA was amplified using specific primers for cox1 gene, i.e., forward JB3 primer (5-TTT TTT GGG CAT CCT GAG GTT TAT-3) and reverse JB4.5 primer (5-TAA AGA AAG AAC ATA ATG AAA ATG-3'] as described by Bowles et al. [5]. PCR was done in a 25 µl final volume containing 10 µl master mixes (Type-it HRM PCR Kit, Qiagen, Hilden, Germany), 9 µl distilled water, 1 µl each primer, and 4 µl template DNA.
Enzymatic reaction was performed under the following conditions: an initial denaturation step at 95℃ for 10 min, followed by 40 cycles at 95℃ for 10 sec, 55℃ for 30 sec, 72℃ for 27 sec, and a final extension step at 72℃ for 5 min.
The master mix provided with the kit contains the novel double-stranded DNA-binding fluorescent dye, Eva Green, and includes an optimized HRM buffer, Hot StarTaq plus DNA polymerase, Q-Solution, and dNTPs. Together, these components provided reliable PCR specificity and acceptable results, even with hard genomic loci. Real-time PCR was carried out in a Mini Opticon real-time PCR detection system (Rotor-Gene 6,000, Hilden, Germany) under the following conditions: after a pre-incubation step (95℃ for 3 min) to activate the polymerase enzyme, 40 cycles of amplification were performed, each one including 95℃ for 10 sec, 55℃ for 30 sec, and 72℃ for 27 sec, followed by a final extension at 72℃ for 5 min. The fluorescence signal was measured at each cycle after the extension step. The melting experiment was performed from 55℃ to 95℃ at 1℃/sec with continuous fluorescence monitoring. Melting peaks were visualized by plotting the first derivate against the melting temperature; the Tm was defined as the peak of the curve. The quantitative detection and Tm were obtained using the Rotor-gene 6,000 series software version 1.7 (Corbett, Hilden, Germany).
Two replicates of each sample were analyzed and Tm analysis was repeated 3 times in each run to confirm the repeatability of the Tm assay by estimating the Tm variation within PCR amplification (intra-assay) and between PCR amplification (inter-assay), respectively. The coefficient of variation (CV) was calculated dividing the standard deviation by the arithmetic mean of the measured values of Tm (CV=standard deviation [SD]/mean value).
In addition, to check the uniformity of temperature of the cycler block, indicating the reliability of results, replicates of an already genotyped sample were amplified in different positions of the cycler block within the same amplification cycle. The intra-assay CVs represent the mean CVs for the results obtained from the replicates of all the E. granulosus genotypes in the same run. The inter-assay CVs represent the mean CVs for the results obtained from 3 separate runs.
To confirm the identified genotypes, 22 samples were randomly sequenced for cox1 mtDNA. Three samples that have been already sequenced for cox1 and identified as G1, G3, and G6 (accession nos. HM563013, HM563017, and HM563018, respectively) were included in each PCR set as positive controls.
Sequence homology and phylogenetic analysis
The obtained continuous sequences were compared with previously published sequences of the mitochondrial cox1 for E. granulosus genotypes in NCBI using BLAST system. Phylogenetic analyses of the sequence data were inferred with maximum likelihood and multiple alignments and done using MEGA 5 of the computer program CLC main workbench software (version 5.2.1, 2013) [24,25,26].
RESULTS
Out of 71 isolates, 5 cattle isolates were sterile and excluded from further investigation. Molecular survey and PCR amplification were successfully performed on all 66 isolates. Of 66 samples, 51 isolates were collected from sheep (35 livers and 16 lungs), 8 from goat (4 livers and 4 lungs), and 7 cattle origin (lungs). Real-time PCR using the partial sequences of cox1 gene of 66 E. granulosus isolates showed that 49, 15, and 2 isolates were identified as G1, G3, and G6 genotypes, respectively. Sheep, goat, and cattle isolates were genotypically categorized as G1 (77%, 63%, and 72%), G3 (21%, 25%, and 28%), and G6 (2%, 12%, and 0%), respectively (data not shown).
As shown in Table 1, real-time PCR melting curve results indicated that the mean Tm of G1, G3, and G6 genotypes were 79.3, 78.8, and 77.3, respectively. Assessment of intra and inter-assay variability showed low and acceptable CVs (Table 1). Melting curve and Tm of the analyzed hydatid cysts identification by sequencing are shown in Fig. 1.

Mean Tm (℃) and SD calculated and intra- and inter-assay CVs of the G1, G3, and G6 genotypes of E. granulosus
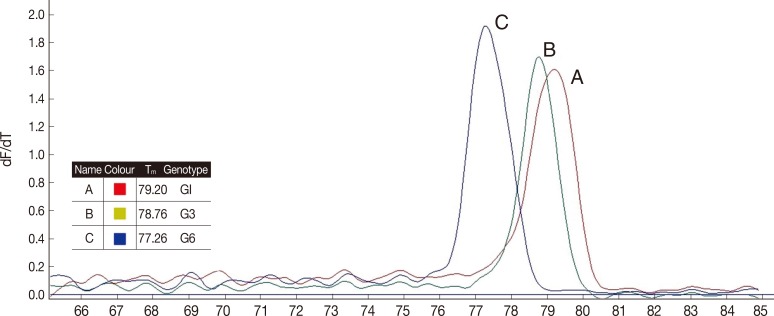
Tm of the analyzed hydatid cysts. (A) E. granulosus G1 identified by sequencing, (B) E. granulosus G3 identified by sequencing, and (C) E. granulosus G6 identification by sequencing.
Sequencing results confirmed that all isolates were correctly differentiated by real-time PCR. All the isolates identified by real-time PCR were clustered along with the corresponding reference genotypes as shown in Fig. 2. The phylogenetic tree was divided into 2 main clades, the first clade contained 2 subclades corresponding to the G1 and G3 genotypes (E. granulosus sensu stricto), and the second one corresponding to the isolates identified as G6 (E. canadensis) along with the individual reference genotypes. Forty-nine samples with Tm between 79.1℃ and 79.5℃ had identical sequences to the G1genotype, while the 15 samples with Tm between 78.6℃ and 79.0℃ had identical sequences to the G3 genotype, and finally 2 samples with Tm between 77.1℃ and 77.5℃ had identical sequences to the G6 genotype.
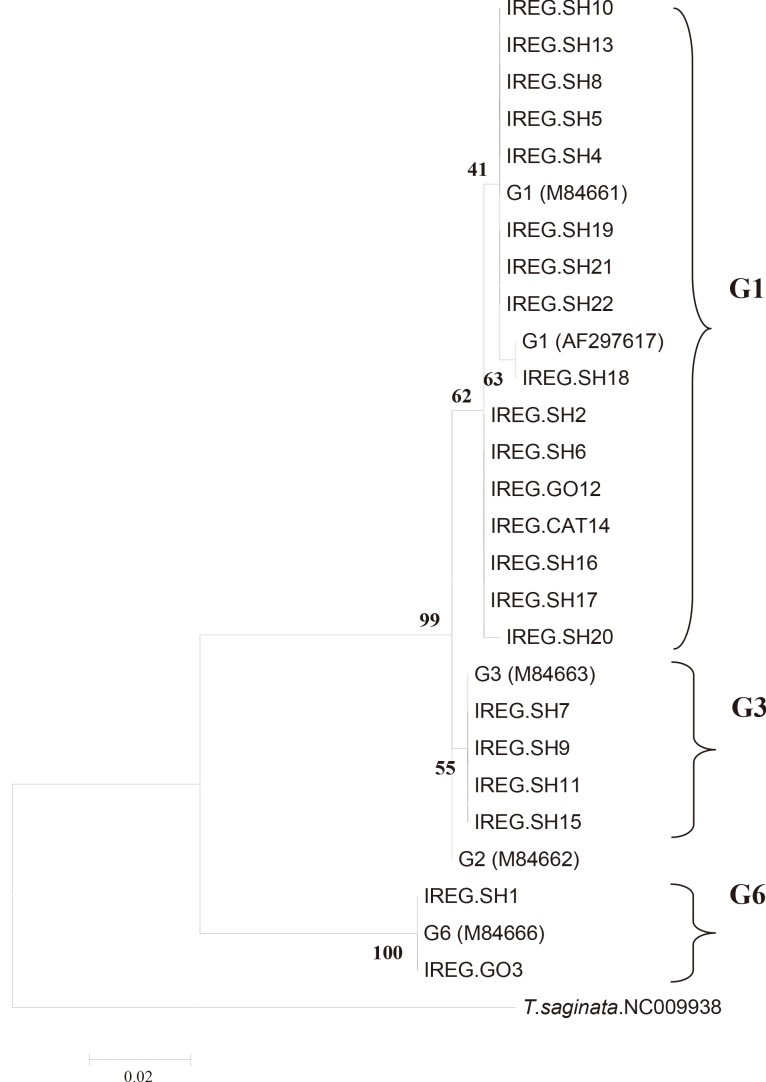
Molecular phylogenetic tree of 22 E. granulosus isolates of sheep, goat, and cow along with reference isolates based on CO1 gene sequence. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2 parameter model [19]. The tree with the highest log likelihood (-814.2241) is shown. Reference accession nos.: G1, M64661; G3, M64663; G6, M84666.
Phylogenetic analysis also showed that 16 isolates of G1 genotype had micro-variants, compared to the original sequences of cox1 gene (accession no. M84661). Four samples identified as G3 and 2 samples identified as G6 were completely identical to the G3 and G6 reference sequences (accession nos. M84663 and M84666, respectively).
DISCUSSION
In the present study, Tm analysis was used for detection of E. granulosus strains (G1, G3, and G6 genotypes) in Isfahan Province, Iran. Due to high sensitivity, specificity, accuracy, and repeatability of this novel method, we confirmed that this technique is useful for detection of Iranian genotypes. Because of 0.5℃ mean difference of Tm between G1 and G3 genotypes, the Tm ranges did not overlap, therefore we could differentiate these 2 genotypes easily and rapidly. On the other hand, G6 genotype could be easily distinguished from the other ones (G1 and G3), because of 1.5℃ different Tm. The accuracy of the Tm assay was confirmed by comparing these results with direct sequencing.
The advantage of using real-time PCR is that this technique is more cost-effective and less time-consuming than conventional PCR. Actually, with this method, a large number of samples could be tested quickly and repeatable results could be achieved with significant saving in time and materials [17,27,28]. On the other hand, this method has some disadvantages, such as existing instruments with limited multiplexing capabilities, incompatibility of several systems with some fluorogenic chemistries, and necessity of high technical skills for its launching [27].
Epidemiological studies showed that G1 genotype is the predominant genotype in both human and animals [29]. Maurelli et al. [17] used real-time PCR for identification of G1 and G2/G3 genotypes of E. granulosus in Italy. They found that Tm analysis can be used for discriminating between G1 and G2/G3 genotypes of E. granulosus sensu stricto. Our results were compatible with their findings.
Recently, Boubaker et al. [30] in Switzerland developed a single-tube multiplex PCR (mPCR) for discrimination of the 3 levels of the E. granulosus complex genotyping. They found E. granulosus sensu stricto (G1/G2/G3), E. granulosus equinus (G4), and E. canadensis (G7) as predominant genotypes in their study. This study showed that this method could provide the unique opportunity to address directly speciation and genotyping within the framework of large-scale studies.
Recent studies conducted analysis of various strains in the Middle East. For example, a study by Adwan et al. [12] in Palestine showed the presence of G1-G3 genotypes in this region using cox1 loci. In another study, Simsek et al. [31] reported a G1/G3 cluster (E. granulosus sensu stricto) in Turkey.
In recent years, several studies reported different genotypes of E. granulosus from intermediate and definitive hosts, including the sheep, goat, cattle, camel, buffalo, human, and dog in different parts of Iran [4,22,32,33]. The results of this study were similar to previous studies in Iran showing that genotypes G1, G3, and G6 are present in intermediate hosts [29]. In the few studies, genotypes G1, G3, and G6 were simultaneously reported [4,34]. Recently, Rostami et al. [29] used the post real-time PCR with HRM method and differentiated G1, G3, and G6 genotypes of sensu lato in Iran. Rajabloo et al. [35] reported G1 and G6 genotypes from goats in Iran. A similar study in Ilam Province on hydatid cysts isolated from animals and humans showed E. granulosus sensu stricto (G1-G3) in this region [36]. A study on 120 hydatid cysts isolated from goats in Mazandaran Province, Northern Iran, by Youssefi et al. [37] showed the G1 genotype as the dominant strain. Among various studies, Rostami et al. [29] reported G1, G3, and G6 genotypes in the intermediate hosts that are similar to our results.
In conclusion, we believe that real-time PCR that has been developed in the present study, provides powerful means for molecular and epidemiological studies on human and animal E. granulosus infections. Therefore, we suggest that the extensive use of this technique with high sample size can help to find undetected genotypes using different molecular targets.
ACKNOWLEDGMENTS
This study was carried out as a part of M.Sc. thesis done by Ahmad Hosseini Safa and was financially supported by the vice-chancellor for Research, Isfahan University of Medical Sciences, Iran.
Notes
The authors declare no conflict of interest related to this work.