Prevalence of Cryptosporidium-Associated Diarrhea in a High Altitude-Community of Saudi Arabia Detected by Conventional and Molecular Methods
Article information
Abstract
Cryptosporidium diarrhea represents a relevant clinical problem in developing countries. In Al-Taif, a city of Saudi Arabia that lies at an altitude of an around 2 km above the sea level, Cryptosporidium infection seems to be undiagnosed in nearly all clinical laboratories. Furthermore, nothing was published regarding Cryptosporidium-associated diarrhea in this area. The objectives of this research were to (1) determine the Cryptosporidium prevalence among patients with diarrhea and (2) to estimate the performances of 3 different diagnostic methods. Total 180 diarrheal fecal samples, 1 sample per patient, were collected between January and August 2013. Samples were screened for Cryptosporidium with modified Zeihl Neelsen (ZN) microscopy, RIDA® Quick lateral flow (LF) immunotest, and a previously published PCR. The Cryptosporidium prevalence rate was 9.4% (17/180), 10% (18/180), and 11.6% (21/180) by microscopy, LF, and PCR test, respectively. Infection was significantly (P=0.004) predominant among children <5 years (22%) followed by children 5-9 years (11.1%). Although infection was higher in males than in females (16.2% males and 8.5% females), the difference was not statistically significant (P=0.11). Compared to PCR, the sensitivity of microscopy and the LF test were 80.9%, 85.7%, respectively. To conclude, high Cryptosporidium-associated diarrhea was found in this area especially in children ≤9 years. The PCR test showed the best performance followed by the LF test and ZN staining microscopy. The primary health care providers in Al-Taif need to be aware of and do testing for this protozoon, particularly for children seen with diarrhea.
INTRODUCTION
Cryptosporidium has been recognized as an important human enteric parasite since 1976 [1]. Many cases of non-bacterial, non-viral infectious human diarrhea are commonly attributed to Cryptosporidium [2]. Cryptosporidium infection can easily occurred by ingestion of ≤10 oocysts contaminated in water or food [3]. These small sized oocysts (4-6 µm) are highly resistance to common household disinfectants and can stay infectious for a long period in the environment [4]. The source of infection could be a human or an animal. All these factors gave Cryptosporidium infection a major public health implication. Cryptosporidiosis, the disease, is often asymptomatic in endemic areas or it can cause diarrhea in others [3,5]. Diarrhea is usually mild and self-limiting in patients with good immunity. However, it could be severe, long-standing, and life threatening in patients with incompetent immunity [6]. This Cryptosporidium-associated diarrhea is not unique. Thus, diagnosis should be relied on laboratory diagnostic tests.
In spite of major advances made in its diagnostic techniques, Cryptosporidium infection lacks the routine diagnosis in most of clinical laboratories [7]. Based on the characteristic morphology of the oocysts, few microscopic techniques are commonly used for Cryptosporidium detection in feces. However, a complex clinical specimen like feces makes identification not an easy task. It is also time-consuming and tedious, and requires a great experience [8]. Certain stains such as acid fast and auramine/rhodamine have facilitated oocyst detection but on the expense of time and costs. To simplify the protozoan detection, many immunoassay-based diagnostic tests became available and frequently used in clinical laboratories. These assays rely on detection of species-specific oocyst surface antigens in feces (i.e., coproantigens). The coproantigen testing is simple, easy to perform, and ideally used to screen a large batch of samples within reasonable time. However, because of antigenic variability within Cryptosporidium clinical isolates, a major sensitivity conflict of these assays over microscopy is reported [9].
Human infection is frequently attributed to at least 2 distinct, human or calf Cryptosporidium isolates. Identification of the isolate is far important to control the spread of infection especially in outbreak situation. Neither microscopy nor coproantigen assays can achieve that purpose. Therefore, in recent years, many PCR-based assays were developed to detect and characterize Cryptosporidium DNA in feces [10]. However, because of its high cost, PCR is still confined to research laboratories and a few specialized reference laboratories.
In Saudi Arabia, few reports have studied the prevalence of Cryptosporidium infection in various populations [11,12,13]. All these populations are living in low or normal altitude regions and all the estimated prevalence rates were based on microscopic methods. In a previous study, we developed an extraction protocol that was capable, with a previously published PCR protocol, to detect ≈2 oocysts per extract using oocysts-spiked specimens. This assay was proved 100% sensitive and specific using Cryptosporidium control samples [14].
In this context, an attempt was primarily made to know the prevalence rate of Cryptosporidium in Al-Taif, a Saudi city lying at an altitude of an around 2 km above the sea level. We also attempted to estimate the diagnostic performance of the widely-studied microscopic method in comparison to a commercially available lateral flow (LF) immunoassay and PCR protocol using clinical samples from this high altitude area.
MATERIALS AND METHODS
Ethical consideration
This research was carried out after approval gained from the Local Directorate of health as well the ethics committee of Taif University, Saudi Arabia.
Samples collection and storage
Fecal specimens were collected from 180 diarrheal patients. Specimens were taken from samples submitted for routine parasitological examinations at 2 public hospitals in Taif city, during the period of January to August 2013. Samples, 1 for each patient, were collected in sterile containers and sent within 2 hr to our Faculty's Laboratory, stored at 4℃ for parasitological examinations.
Microscopic diagnosis
An aliquot from each specimen was fixed with 10% formalin for half an hour and then concentrated by formol-ether sedimentation method according to Humphries et al. [8]. A moderately thick, wet mount smear was prepared and permanently stained following the modified Zeihl Neelsen (ZN) staining procedure [15]. Each smear was thoroughly examined by a binocular light microscopy with the ×40 and ×100 objective lenses.
Immunodiagnosis
The Cryptosporidium oocysts surface antigen found in feces (i.e., coproantigen) were searched for in fresh unpreserved samples using RIDA® Quick Cryptosporidium LF test (R-Biopharm, Darmstadt, Germany). The test was carried out following the kit manufacturer's protocol.
Molecular diagnosis
Fecal aliquots without any additive were subjected to PCR test within 2 weeks from storage. DNA samples were isolated and purified using QIAamp® Stool Mini Kit (Qiagen, Hilden, Germany). The extraction procedure was carried out following the changes we have introduced over amended kit's protocol in a previous study [14]. A specific DNA sequence of the Cryptosporidium oocyst wall protein (COWP) gene was amplified using previously published primers [16]. Amplification reactions were done using Techne™ TC-4000 thermal cycler. The GoTaq® Hot Start Polymerase (Promega, Heidelberg, Germany) and other reagents were used in PCR with final concentrations closely similar to the published protocol. PCR products were analyzed on 1-2% of agarose gel electrophoresis.
Data collection, processing, and analysis
Tests results of clinical samples together with the clinical data obtained by revising the corresponding laboratory reports were analyzed through the Statistical Package for Social Science (SPSS) version 17. Chi-square test was adopted to test associations between age groups, gender, and Cryptosporidium positivity variables. However, if any expected value was less than 5, Fisher's exact chi-square test was used. Differences in data were considered statistically significant if the P-value was below 0.05. The diagnostic sensitivity (SE), specificity (SP), positive predictive value (PPV), and negative predictive value (NPV) of various diagnostic assays were calculated by taking the PCR test results as the gold standard.
RESULTS
Prevalence by ZN staining microscopy
Of all 180 specimens screened by microscopy, Cryptosporidium oocysts (Fig. 1) were seen in 17 wet mount preparations stained with modified ZN method. The estimated prevalence rate was 9.4% (17/180; Table 1).
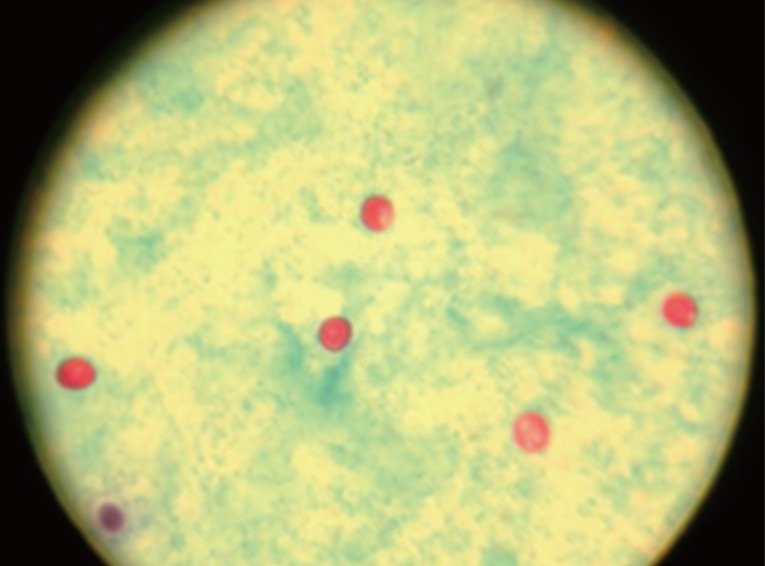
Microscopic picture of Cryptosporidium oocysts (red) in a wet mount smear prepared from a fecal specimen concentrated by modified formol-ether method and stained following the modified Zeihl Neelsen procedure (×400 magnification).
Prevalence by the LF test
The Cryptosporidium coproantigen was identified in 18 specimens with a prevalence rate of 18/180 (10%). All samples diagnosed as Cryptosporidium-positives by microscopy were also positives by the LF test. One sample diagnosed as Cryptosporidium-negative by microscopy was positive by the coproantigen detection test (Table 1). This sample was from a 2-year old child and proved to be positive also by the PCR test.
Prevalence by the PCR assay
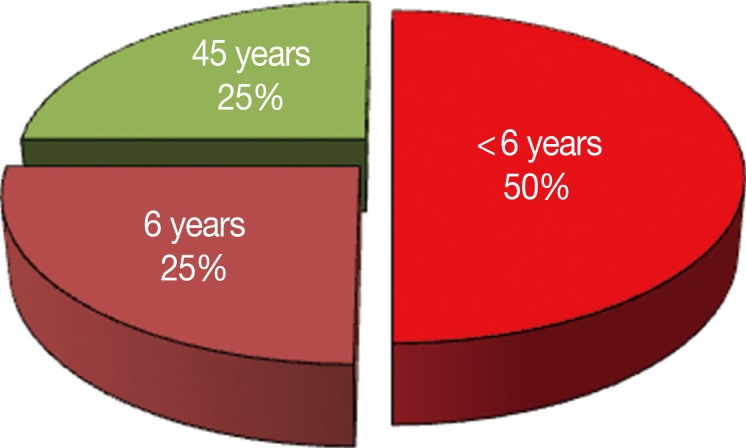
The fecally recovered Cryptosporidium target DNA was successfully identified in 21 samples by PCR. Accordingly, the Cryptosporidium prevalence rate was 11.6% (21/180). All the Cryptosporidium-positive specimens by microscopy and LF test were also positive by the PCR test. The PCR test picked up 4 positive samples more than microscopy (Table 2) and 3 positive samples more than the LF test (Fig. 2; Table 3). These samples were from 2 children under 5 years of age, 1 child aged 6 years and a patient aged 45 years (Fig. 3).
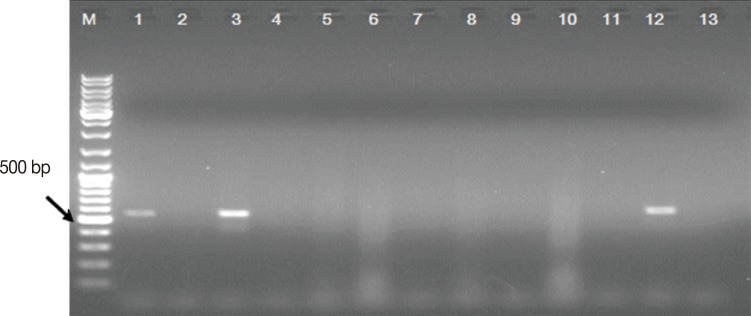
Representative ethidium bromide-stained 1% agarose gel electrophoresis picture showing the PCR test results of 12 samples (Lane 1-12) diagnosed as Cryptosporidium-negative by the LF test. M: GeneRuler™ 100 bp DNA marker; Lanes 1, 3, and 12: PCR products, ≈550 bp each, recovered from Cryptosporidium DNA-positive samples. Lane 2, 4-11: Cryptosporidium DNA-negative samples. Lane 13: PCR-negative control (master mix without template DNA).
Prevalence as distributed by age and gender
As seen in Table 4, the predominant infection was among children <5 years (22%, 11/50) followed by children 5-9 years (11.1%, 5/45). There was a significant difference between different age groups and Cryptosporidium infection (P=0.004). Of the 21 Cryptosporidium-positive samples by PCR, 12 were obtained from males (16.2%, 12/74) while the remaining 9 were from female patients (8.5%, 9/106; Table 5). However, the difference was not statistically significant (P=0.11).
Diagnostic performance of microscopy and LF test
Taking the PCR test results as the gold standard, the modified ZN-stained microscopy showed SE, SP, NPV, PPV of 81.0%, 100%, 97.6%, and 100%, respectively (Table 6). By the same principle, the LF test showed SE, SP, NPV, PPV of 85.7%, 100%, 98.2%, and 100%, respectively (Table 6).
DISCUSSION
Unlike other studies carried out in Saudi Arabia and focusing in high risk groups for Cryptosporidium infection such as children or patients with human immunodeficiency virus (HIV), we tried, in the current research work, to estimate the prevalence of Cryptosporidium infection in patients of different ages seen with diarrhea. Our study could be biased by focusing on the diarrheal specimens that carry a higher possibility of being Cryptosporidium-positive than that would be found in a routine diagnostic laboratory. However, this selection bias was not taken great considerations because of the followings: First, although very little data is available for the current diarrhea incidence estimates in Al-Taif, a relatively old estimate of 25% has been previously reported [17]. To highlight the importance of Cryptosporidium to the primary health care providers, as an important etiology of the infectious diarrhea in the study population, these diarrheal specimens were chosen. Second, most of the previous survey studies have used diarrheal specimens to estimate the prevalence rates in Saudi areas of various altitudes and this make comparison among these studies wiser. Last, nearly all the clinical laboratories at Al-Taif do not routinely test for Cryptosporidium unless requested by physicians. Therefore, it was important, in the present study, to estimate the prevalence rate of Cryptosporidium using diarrheal speicmens from this community.
To the best of our knowledge, this study is the first that interested to estimate the prevalence of Cryptosporidium in a high-altitude like Al-Taif region. The high-altitude area does not house specific infectious agents by itself. However, it carries environmental stressors such as increased ultraviolet radiation, hypobaria, hypoxemia, uncertain climate, and shortage of acceptable drinking water. Pathogens, including Cryptosporidium, may be affected by these stressors, but nothing was published about their possible role [18]. In this high-altitude community, Cryptosporidium infection was identified with estimated prevalence of ≈10%, a rate that has not been reached in other Saudi regions with different altitudes. In Riyadh, a low-altitude area that lies at 620 m above the sea level, a prevalence rate of ≈8% (11/136) was reported among high risk HIV-positive patients [19]. Lower rates (1-4%) were shown among children seen with diarrhea from areas lying at the sea level like Dammam, Al Khobar, Jeddah, and Makah [11,12,13]. The high prevalence of Cryptosporidium infection could be attributed to the shortage of drinking water resources that is common in this region. However, until further in-depth studies undertaken, the risks of high-altitude for Cryptosporidium infection can not be fully postulated.
Although all age groups can get the infection, the greatest number of Cryptosporidium-positive samples, in this study, was for children. Also, it was shown that the younger age represent a risk factor for developing Cryptosporidium-associated diarrhea. Specimens collected from children <5 years of age were 2 times more likely to be infected with Cryptosporidium as compared to those collected from elder children. Our findings are consistent with many previous studies carried out in Saudi populations [11,12,13,19] or other populations [20,21,22]. The gender have not been considered as an actual risk for Cryptosporidium infection in many previous reports [11,12,13,19], although elsewhere, higher infection in females than in males has been reported [23,24]. Interestingly, in this study, Cryptosporidium infection was higher in males than females. This could be explained by the fact that males, especially in this community, have more freedom than females to go outdoors and practice activities such as dealing with farm animals, drinking unprotected underground well-water, and swimming in public pools.
Because of the little information included in the laboratory request form data, other risk factors such as the patient immune status, residence and outdoor activities, were not investigated in this study.
Based on the single stool examination, ZN staining microscopy demonstrated the lowest sensitivity among the 3 tests used in the study. The sensitivity of the technique can be increased with increasing the number of samples to 3 taken in consecutive days. Similar to the other 2 diagnostic tests used in the current study, ZN staining microscopy proved very specific. Also, the rapid immunoassay test used in the current study proved to be very useful for identification of the Cryptosporidium surface antigen in feces. It picked 1 more positive sample than microscopy. This sample could be for a patient recovering from an infection or could be due to sensitivity variations. In the first case, the oocyst antigens shed in the absence of intact oocysts in feces, while in the second case, the oocysts shed in feces in a number below the detection limit of microscopy. In a previous work [25], we reported a sensitivity of 95% for the same kit but as part of a 3 kits-based screening algorithm. Also, a lower sensitivity of the same kit was displayed in a previous study [26].
Our results showed that the PCR test together with the extraction protocol was proved to be the most sensitive technique among the 3 kinds used in the study, a finding that was consistent with previous studies [27,28,29]. Although the target cowp gene DNA sequence adopted in the PCR assay permits identification of the most common human species and genotypes through restriction digestion or direct sequencing analysis of the PCR products, no attempt was made to characterize the Cryptosporidium-positive samples in this study due to shortage of funding this research. Much is still to be learned about the epidemiology of this protozoan infection in this high-altitude region and has to be investigated in the future studies.
To conclude, our study has shown that Cryptosporidium infection is prevalent in the community studied. The infection rate was more prevalent in this high altitude area (≈ 10%) than what has been estimated in areas lying at low-altitude or at the sea level. However, these correlations require further in-depth studies to be proved. Infections were more in children than elder patients and in males than in females. Also in the current study, the PCR test demonstrated the best performance among the 3 tests used. To ensure that Cryptosporidium infection does not go undiagnosed, 1 of the 3 tests could be used for the diagnosis especially in children seen with diarrhea.
ACKNOWLEDGMENTS
We are indebted to all laboratory personnel at the microbiology departments at king Abdul-Aziz specialized hospital, King Faisal hospital, and the specialized children hospitals for their great help during the study. Special thanks go to Mr. Alsalmani H. and Mr. Althobati K. (final-year students at the College of Applied Medical Sciences, Taif University) for their help in collecting clinical samples and patients' data.
Notes
We have no conflict of interest related to this work.