Occurrence and Molecular Identification of Anisakis Dujardin, 1845 from Marine Fish in Southern Makassar Strait, Indonesia
Article information
Abstract
Anisakis spp. (Nematoda: Anisakidae) parasitize a wide range of marine animals, mammals serving as the definitive host and different fish species as intermediate or paratenic hosts. In this study, 18 fish species were investigated for Anisakis infection. Katsuwonus pelamis, Euthynnus affinis, Caranx sp., and Auxis thazard were infected with high prevalence of Anisakis type I, while Cephalopholis cyanostigma and Rastrelliger kanagurta revealed low prevalence. The mean intensity of Anisakis larvae in K. pelamis and A. thazard was 49.7 and 5.6, respectively. A total of 73 Anisakis type I larvae collected from K. pelamis and A. thazard were all identified as Anisakis typica by PCR-RFLP analysis. Five specimens of Anisakis from K. pelamis and 15 specimens from A. thazard were sequenced using ITS1-5.8S-ITS2 region and 6 specimens from A. thazard and 4 specimens from K. pelamis were sequenced in mtDNA cox2 region. Alignments of the samples in the ITS region showed 2 patterns of nucleotides. The first pattern (genotype) of Anisakis from A. thazard had 100% similarity with adult A. typica from dolphins from USA, whereas the second genotype from A. thazard and K. pelamis had 4 base pairs different in ITS1 region with adult A. typica from USA. In the mtDNA cox2 regions, Anisakis type I specimens from A. thazard and K. pelamis showed similarity range from 94% to 99% with A. typica AB517571/DQ116427. The difference of 4 bp nucleotides in ITS1 regions and divergence into 2 subgroups in mtDNA cox2 indicating the existence of A. typica sibling species in the Makassar Strait.
INTRODUCTION
The Indonesian archipelago, which comprises of more than 17 thousands islands and coastal lines length of about 104,000 km, provides great natural resources for Indonesia economic growth in many regions, particularly the fishery sector. Total production from fisheries continues to grow from 8.2 million ton in 2007 to 12.4 million ton in 2011 [1]. Indonesia is also recognized as a center of marine organism diversity and is inhabited with abundant fish species. Fishery products, which are either traded as export commodities or for local consumption, have to be free from any zoonotic parasites, such as anisakid nematodes. Food safety has recently become a great concern for consumers. Zoonotic parasites in fish products are mainly caused by helminths, which utilize fish as the intermediate host, and marine mammals such as dolphins or whales as the final host [2-4]. Some helminths are transmittable and able to survive in the human digestive tract after consuming raw fish infected by larval helminths, causing significant clinical diseases, as well as allergic reactions [5-7]. Symptoms of anisakiasis include epigastric pain, nausea, vomiting, and diarrhea [8]. Among the helminth parasites, larval nematodes of the genus Anisakis are commonly found in the musculature or digestive tracts of many species of marine fish. Anisakis simplex is the most well-known zoonotic nematode which has been reported to cause anisakiasis of humans in many countries in Europe and Asia [8]. Though reported cases of anisakiasis in Indonesia are very rare, a study conducted by Uga et al. [9] using a seroepidemiological approach of inhabitants in East Java revealed that about 11% of samples were positive for Anisakis infection.
In previous studies, Anisakis spp. could only be categorized morphologically into Anisakis type I and type II, in which the former has a longer ventriculus and a mucron, while the latter has short ventriculus and no mucron [10]. Identification to the species level by microscopic examinations is usually unreliable due to undeveloped morphological characteristics of larval stage nematodes. However, accurate identification of Anisakis nematodes larvae is required for precise diagnosis of Anisakis infections in humans and fish and to improve food safety. In addition, they can be used as biological indicators in the study of stock discrimination of migratory fish [11-15]. Recent studies showed that molecular diagnostic techniques could be used to identify Anisakis to a species level. PCR-RFLP has been widely used to identify Anisakis spp. in different fish species [16-20]. Molecularly, the previous Anisakis type I is known to consist of 6 species (A. ziphidarum, A. nascettii, A. typica, and 3 sibling species of A. simplex complex, namely A. simplex (sensu stricto) (s.s.), A. pegreffii, and A. simplex C, whereas type II consists of 3 species (A. paggiae, A. brevispiculata, and A. physeteris) [21,22]. Recent studies in Japan showed that L3, L4, and adult of A. simplex (s.s) and A. pegreffii could be distinguished morphologically based on the ventricululs length, in which the former has a longer (0.90-1.50 mm) ventriculus than the latter (0.50-0.78 mm) [23].
Parasitological research on Anisakis spp. in Indonesia is relatively scarce. Research on Anisakis was previously reported from Seribu Islands, Jakarta from 3 species of fish: Rastrelliger kanagurta, Decapterus russelii, and Sardinella sirm [24]. The study has found 2 types of Anisakis, i.e., Anisakis type I and Terranova type B, where Anisakis type I predominated. A recent study conducted on several fish species of Balinese and Javanese waters has found 3 species of Anisakis; A. typica, Anisakis sp. 1, and Anisakis sp. 2 [25]. Another study of marine fish in Indonesia in Southern Coast of Kulon Progo, Yogyakarta, has also found 5 out of 11 fish species examined harbored Anisakis spp. [26]. However, from most studies conducted in Indonesia, except for that conducted by Palm et al. [25], identification of the Anisakis larva was solely based on morphology, making it unreliable for species identification.
The aim of the present study was to investigate the occurrence of Anisakis infection from some marine fish in the Southern Makassar Strait, and characterize them to species level using PCR-RFLP genetic analysis, the molecular keys described by D'Amelio et al. [27] and Pontes et al. [17], and sequencing of ITS-5.8S and mitochondrial cytochrome c oxidase subunit II (mtDNA cox2) regions.
MATERIALS AND METHODS
Parasitological examinations
A total of 220 fish representing 18 species and 10 families were investigated for Anisakis larvae infection. Fish species, total length, and number of fish examined are given in Table 1. Fish were purchased from local markets located in Makassar, Takalar, and Barru regencies (Fig. 1). The fish were transported to the Laboratory of Fish Parasites and Diseases, Hasanuddin University for parasitological examinations. The length of fish was measured, body cavity was opened, and internal organs were placed on Petri dishes and examined for the presence of Anisakis larvae. Anisakis larvae were distinguished from other anisakids following of the identification protocols of Anderson [28]. Anisakis spp. were then categorized into type I or type II. Anisakis type I was characterized by the presence of boring tooth at the anterior end, ventriculus, and mucron at the posterior end, whereas type II has a boring tooth, ventriculus, and no mucron at the posterior end. Anisakid larvae were isolated from the visceral surface and body cavity of the fish. The larvae were observed under light and dissection microscope for morphological identification. All Anisakis larvae found were counted. Since the number of Anisakis larvae found from fish other than skipjack and frigate tuna was very few (mostly uninfected), Anisakis spp. from these fish were only morphologically identified into Anisakis type I or type II. Only larval Anisakis spp. isolated from skipjack tuna and frigate tuna were used for further molecular characterization.
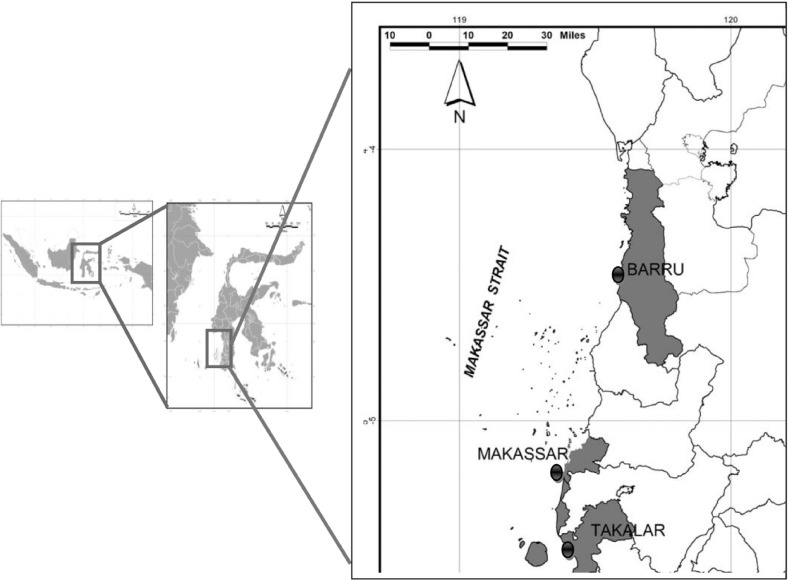
Sampling sites of marine fish at the Southern Makassar Strait: Barru, Makassar, and Takalar Regencies, Indonesia.
The prevalence is defined as the percentage of fish infected by Anisakis larvae. The mean intensity is defined as the total number of Anisakis species found divided by the total number of fish infected [29].
DNA isolation and amplification
Anisakis spp. were fixed and stored in 70% ethanol. Genomic DNA from individual worms was extracted using a QIAamp DNA Mini Kit (Qiagen Inc., Hilden, Germany) following the manufacturer's tissue protocol. DNA was eluted with milliQ and stored at -20℃ before subsequent PCR amplification. Internal transcribed spacer (ITS) and mitochondrial cytochrome c oxidase subunit II (mtDNA cox2) regions were used for amplification and sequencing. The entire rDNA regions comprising ITS1, 5.8S, and ITS2 were amplified using previously described primers NC5 (5'-GTAGGTGAACCTGCGGAAGGATCATT-3') and NC2 (5'-TTAGTTTCTTTTCCTCCGCT-3') [30]. All PCR was performed in 20 µl which contain approximately dNTP 0.2 mM, primers 0.8 µM, Taq polymerase 0.02 U/µl and 10 x buffer PCR 1X, and 2 µl samples. Milli-Q was added to achieve the total PCR volume. Each PCR reaction was performed in a thermocycler iCycler (Bio-Rad, Hercules, California, USA) under the following conditions: after initial denaturation at 95℃ for 15 min, 30 cycles of 94℃ for 1 min (denaturation), 55℃ for 1 min (annealing), 72℃ for 1 min (extension), followed by a final extension at 72℃ for 5 min. The mtDNA cox2 gene was amplified using the primers 210 (5'-CACCAACTCTTAAAATTATC-3') and 211 (5'-TTTCTAGTTATATAGATTGRT-TYAT-3') [31]. The PCR mixture was denatured at 94℃ for 3 min, followed by 35 cycles at 94℃ for 30 sec, 46℃ for 1 min, 72℃ for 1 min and 30 sec, followed by post-amplification at 72℃ for 10 min. The PCR products obtained were visualized in an SYBR green stained 1.5% agarose gel.
PCR-RFLP
PCR products of about 965 bp amplified with primers NC5 and NC2 were used for PCR-RFLP analysis to identify Anisakis spp. following D'Amelio et al. [27] and Pontes et al. [17]. Three individual restriction enzymes (Taq I, Hinf I, and Cfo I) were used. The PCR products were digested following the manufacturer's recommendation. Briefly, amplicons of 8 µl were mixed with 10 x reaction buffer, 0.5% BSA (only for Taq I), and digested with restriction enzymes Taq I (10 U/µl, Takara) at 65℃ for 3-4 hr, and with Hinf I (10 U/µl, Roche) and Cfo I (10 U/µl, Roche) at 37℃ for 3-4 hr. Milli Q was added to reach a final volume of 20 µl. The digested samples were then separated by electrophoresis using 1.5% agarose gel at 100 V for 40 min, stained with SYBR green, and photographed. Their size was estimated using 100 bp ladder marker (Takara).
Sequencing
Twenty specimens, 5 from K. pelamis and 15 from A. thazard were molecularly identified using PCR-sequencing in ITS1-5.8S-ITS2. In addition, 10 Anisakis samples (6 from A. thazard and 4 from K. pelamis) were sequenced in mtDNA cox2 region. PCR products were purified using a PCR purification kit (Qiagen) and used directly in sequencing reactions. A 100 bp or 1 kb ladder marker (Takara) was used to estimate the size of PCR products. Afterwards, a total volume of 14 µl containing 6.4 pmol primer and 10 to 40 ng DNA was prepared and sent to Operon Biotechnologies Company (Tokyo, Japan) for sequencing. Milli-Q was added when necessary for DNA dilution to meet the concentration of DNA required. Both spacers (ITS1 and ITS2) and the 5.8S gene were sequenced in both directions from each PCR product, using the same primers as above (NC5 and NC2), NC13 (forward; 5'-ATCGATGAAGAACGCAGC-3'), NC13R (reverse; 5'-GCTGCGTTCTTCATCGAT-3'), and XZ1R (reverse; 5'-GGAATGAACCCGATGGCGCAAT-3'). Both forward (primer 210) and reverse (primer 211) directions of mtDNA cox2 region was sequenced using the same primer as used for PCR amplification.
Alignment and phylogenetic analysis
The forward and reverse sequences of ITS (ITS1, 5.8S, and ITS2) and mtDNA cox2 regions were assembled and edited using Bioedit Allignment Sequence Editor Ver. 7.0.5.3. They were compared manually with the original chromatograms when necessary. The obtained sequences were aligned with previously characterized sequences of Anisakis spp. registered in GenBank, using CLUSTAL X Version 2.1 Multiple Sequence Alignments [32]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 [33]. Maximum likelihood tree was constructed for ITS-5.8S using Pseudoterranova decipiens as an outgroup and neighbour-joining tree for mtDNA cox2 using H. reliquens as an outgroup. ITS-5.8S and mtDNA cox2 gene sequences were deposited in GenBank under accession no. KC928261 to KC928272.
RESULTS
Prevalence and intensity of Anisakis larvae
Morphologically, all Anisakis larvae found were identified as Anisakis type I. Among the fish examined, K. pelamis, E. affinis, Caranx sp., and A. thazard were infected at a high prevalence and high intensity, while other fish (C. cyanostigma and R. kanagurta) were infected with a low prevalence and low intensity (Table 1). The remainders were not infected. The parasites were mainly found on the surface of the viscera such as the liver and intestines, and no Anisakis larvae were found in the muscle. For identification, the parasites were cleaned and the sheath removed. Anisakis larvae can be distinguished from other anisakid larvae such as members of Pseudoterranova, Hysterothylacium, and Contracaecum based on the shape of the ventriculus, which is clearly visible under a stereomicroscope. The prevalence of Anisakis type I in K. pelamis was 92.3%, and mean intensity was 49.7 parasites/fish. The highest number was 175 parasites in an individual fish. The prevalence of Anisakis type I larvae in A. thazard (33-41 cm in total length) reached 46.7% with a mean intensity of infection of 5.6 parasites/fish. However, none of A. thazard of smaller size (19-25 cm in total length) were infected out of 12 fish examined. The other fish with a high prevalence of Anisakis type I were Caranx sp. (75%) and E. affinis (66.7%) (Table 1).
PCR-RFLP patterns
Amplification of entire ITS and 5.8S regions of all specimens of Anisakis produced a PCR product of about 960 bp. The PCR products were digested using 3 different restriction enzymes, Taq I, Hinf I, and Cfo I. In PCR-RFLP, all specimens digested with Taq I, Hinf I, and Cfo I indicated that the samples belong to A. typica (Table 2). Based on the RFLP analyses of 73 Anisakis type I specimens from K. pelamis (40 specimens) and A. thazard (33 specimens) were all (100%) identified as A. typica (Table 2).
Sequencing of entire ITS region and mtDNA cox2
Sequencing of entire ITS and 5.8 S regions was performed for 5 samples of Anisakis type I from K. pelamis and 15 from A. thazard. Using the primer in the ITS and 5.8S region approximately 950 bp nucleotides were generated. Nucleotide sequences from all specimens were analyzed using the software Bioedit, and were manually compared with chromatogram when necessary. No variation in the nucleotide sequences was found in Anisakis from K. pelamis, whereas samples from A. thazard showed 2 nucleotide sequence patterns. They differed in 4 base pairs in ITS1 region. The first nucleotide pattern was only recorded from A. thazard, whereas the second one was found from both K. pelamis and A. thazard. The first pattern (genotype) showed 100% similarity with adult A. typica reported from dolphins in USA and high similarity with the Brazilian A. typica from dolphins, which only differed in 3 deletions in ITS1. Whereas the second pattern (genotype) has 4 base pairs difference in ITS1 with A. typica from USA, but 100% similarity with Indonesian A. typica EU346093 from fish Auxis rochei rochei, 4 base pairs different with A. typica EU346092 and 2 base pairs different with A. typica EU346091. A phylogenetic tree using maximum likelihood showed that A. typica found in the present study were in the same clade with other A. typica published in GenBank (Figs. 2, 3). Sequences of 10 samples of Anisakis using the primer in mtDNA cox2 region produced about 600 bp nucleotides. Pair distances of the alignment of mtDNA cox2 showed 94-99% similarity of present samples with A. typica AB517571 from Scomber japonicus, 93-98% with A. typica AB517572 from S. japonicus, and 94-100% with adult A. typica DQ116427 from dolphins (Table 3). The phylogenetic tree of mtDNA cox2 region showed that all samples were in the same cluster with A. typica but produced broad divergence consisting of 2 subgroups (Fig. 4). The first subgroup showed 96% to 100% similarity with the known A. typica, whereas the second one has 93-95% similarity with the nematode (Table 3).

Comparison of ITS1 sequences of Anisakis typica larvae in the present study. KC928261 A. typica (I), KC928262 A. typica (II), and A. typica larva from the bullet tuna from western Indonesia (GenBank accession no. EU346093, EU346092, and EU346091, respectively) were compared with adult specimens from dolphins from Brazil (AY826724) and USA (AB479120). Square shows differences of nucleotides in the ITS1 regions among A. typica.
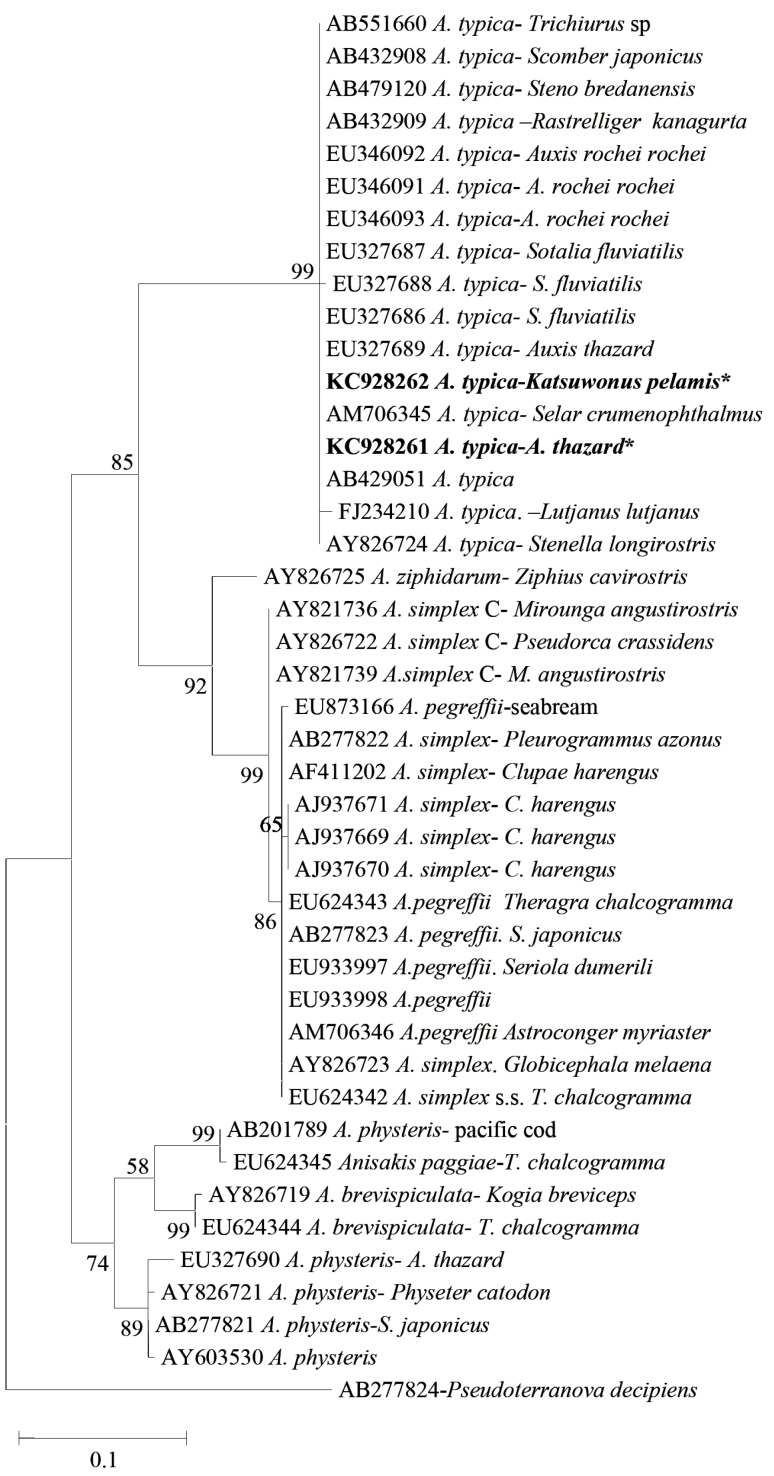
Phylogenetic tree of Anisakis spp. including the present samples (KC928262 A. typica from Katsuwonus pelamis and KC928261 A. typica from Auxis thazard) based on ITS-5.8S-ITS2 gene sequences. Asterisks represent present samples. Maximum likelihood tree was constructed using MEGA version 5.1 [33], drawn using Nearest-Neighbor-Interchange (NNI) with Kimura 2-parameter model and 1,000 boostrap number with complete deletion. Percentages ≥50% are shown at the internal nodes.

Similarity of nucleotide sequences among A. typica including the present Anisakis based on mtDNA cox2 region
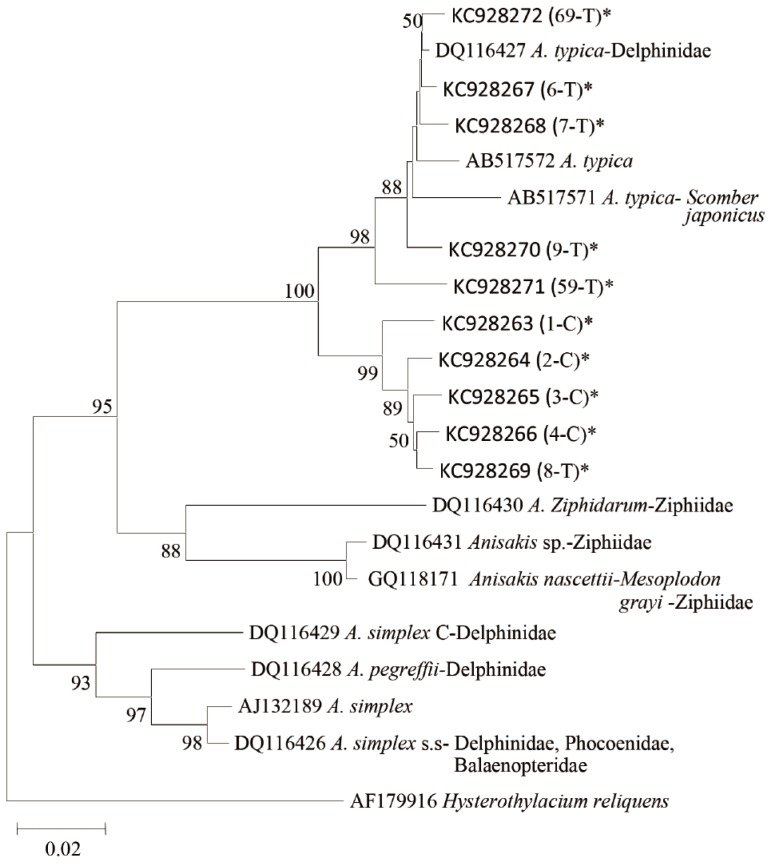
Phylogenetic tree of Anisakis species from the present study (KC928263 to KC928272) and other Anisakis spp. based on mtDNA cox2 gene sequences. Asterisks represent present samples. Neighbour joining tree was constructed using MEGA version 5.1 [33], drawn using Maximum composite likelihood Model and 1,000 boostrap number with complete deletion. Percentages ≥50% are shown at the internal nodes. Sample codes were presented in Table 3.
DISCUSSION
The present study provides molecular identification of Anisakis from K. pelamis and A. thazard using PCR-RFLP and sequencing of ITS-5.8S and mtDNA cox2 regions. This is the first record of molecular identification of Anisakis type I from fish of eastern part of Indonesia. The first molecular identification of Anisakis, which consisted of 3 different genotypes, namely, A. typica, Anisakis sp. 1, and Anisakis sp. 2, was reported from Balinese and Javanese waters [25]. In the present study, based on PCR-RFLP and sequencing, Anisakis larvae were identified as A. typica. PCR-RFLP was further used to detect proportion of Anisakis spp. in fish, and the results showed that from 73 Anisakis type I specimens collected from K. pelamis (40 specimens) and A. thazard (33 specimens), 100% belonged to A. typica. From 20 samples of Anisakis type I sequenced, 2 different patterns, or genotypes, were noted and further identified as A. typica. The 2 patterns differed in 4 base pairs in the ITS1 region. The first pattern showed 100% similarity with adult A. typica (AB479120) reported from dolphins in USA and high similarity with the Brazilian A. typica (AY826724) from dolphins, whereas the second genotype has 4 base pairs different in ITS1 with A. typica from USA (AB479120), and 100% similarity with Indonesian A. typica EU346093 from Auxis rochei rochei. The first genotype was only found in A. thazard, whereas the second one was found in both K. pelamis and A. thazard.
Palm et al. [25] proposed that difference in 4 bp of nucleotide in ITS1 region may indicate the occurrence of A. typica sibling species. Therefore, the present finding suggests that A. typica sibling species may occur in K. pelamis and A. thazard. Phylogenetic trees from the mtDNA cox2 region also showed a cluster within A. typica, and 2 subgroups were noted. The first subgroup consists of all samples from A. thazard and A. typica DQ116427, AB517571, and AB517572. The second subgroup consists of 4 samples from K. pelamis and 1 sample from A. thazard. The first subgroup of the present samples showed 96-100% similarity with the known A. typica, while the second subgroup had 93-95% similarity. The second subgroup forms another cluster which separate them from the known A. typica and this may also indicate the presence of A. typica sibling species. A previous study in Papua New Guinean waters also showed a similar pattern with the present study in which genetic divergence occurred within A. typica clade [34]. Though a study in other nematode taxa showed that sequence differences of about 10-20% were interspecific, and differences of about 7% were regarded as conspecific [35], the present study, as proposed by Palm et al. [25], indicates the presence of A. typica sibling species in K. pelamis and A. thazard, whereas A. typica was recorded only from A. thazard.
Molecular differentiation of Anisakis spp. using PCR-RFLP has been successfully used [16-20,27,35-38]. In the present study, digestion of 73 samples with restriction enzyme Taq I, Hinf I, and Cfo I indicated that the samples were A. typica, and suggested a predominance of A. typica in the eastern parts of Indonesia. This high abundance of A. typica in the present study, and the report of A. typica in the previous study in Balinese and Javanese waters [25], as well as a recent report from Papua New Guinea [34] support the previous findings that this species was more abundant in temperate and tropical waters [39]. Mattiucci et al. [39] recorded larvae of A. typica from A. thazard and Thunnus thynnus from Brazilian Atlantic Ocean, Scomber japonicus and Trachurus picturatus from Madeira Atlantic Ocean, E. affinis, S. commerson, Sarda orientalis, and Coryphaena hippurus from Somali Coast, and Merluccius merluccius from the Mediteranian Sea off Cyprus and off Crete. In Indonesia, more than 34 species of marine fish have been reported to harbor Anisakis spp. [25]. Recent reports of Anisakis infection added 3 more new fish genera to harbor Anisakis spp. Two genera were reported from the Southern coast of Kulon Progo; Parupeneus sp. (Mullidae) and Terapon jarbua (Terapontidae) [26], and 1 genus in the present study, C. cyanostigma (Serranidae), is a new record of Anisakis infection. At present, based on molecular studies, 9 species of Anisakis are known, namely A. simplex s. s., A. pegreffii, A. simplex C, A. typica, A. ziphidarum, A. nascettii, A. physeteris, A. brevispiculata, and A. paggiae [21,22].
A. typica has been reported from numerous marine fish worldwide. The parasite was reported from marine fish in Korea, Japan, China, Portugal, Taiwan, Brazil, Western Indonesia, Morocco, Papua New Guinea, Adriatic Sea of Croatia, Mauritania, and some countries at Mediterranean Sea [20,25,34-37, 40-47]. The existence of A. typica from the Portuguese coast may have extended the distribution of this parasite to cold water. However, the infection level of the parasite was very low. Therefore, it might be possible that the fish may accidently infected through the food chain originating from warm waters. Marques et al. [45] stated that the Portuguese coast is a transition between North-Eastern Atlantic warmer temperate and cold temperate regions so that it might provide an area of species overlap and hence could promote accidental infection.
High prevalence of Anisakis was found in migratory fish, K. pelamis and A. thazard. High prevalence of Anisakis infection was also reported from some marine fish in southern coast of Kulon Progo, Yogyakarta [26]. The prevalence of Anisakis sp. infection was generally higher in bigger fish than in smaller ones [19,25,48]. The same result was found in this study that small A. thazard were not infected with Anisakis, while bigger ones were infected with the prevalence of 47% and the mean intensity of 5.6. This result might be caused by accumulation of the parasites in the big fish due to a long period of infection. Previous reports on Anisakis infection in Indonesian waters showed high prevalence of infection with the parasite in some species of marine fish. Hadidjaja et al. [24] reported that the prevalence of Anisakis type I larvae in Rastrelliger kanagurta, Decapterus russeli, and Sardinella sirm was 49.3%, 50.3%, and 40.9%, respectively, whereas in the present study no Anisakis infection was found in D. russeli and only 5% infection in R. kanagurta. The difference in the prevalence of infection was also noticed at different locations by Palm et al. [25], and they suggested that the high prevalence of Anisakis infection at Northern Balinese coast was due to the high abundance of dolphins, as the final host for Anisakis, in that area. A previous study on the ecology of Pseudoterranova decipiens in Antarctic waters showed that a high prevalence of infection was in accordance with a high abundance of final hosts as well as intermediate hosts in the area [49].
Anisakiasis has been reported from several countries such as Japan, Korea, and some countries in Europe. Anisakis may infect humans and causes anisakiasis after consuming raw infected fish or other marine organisms that function as intermediate hosts. The first report of anisakiasis was from a patient in the Netherlands who had gastrointestinal problems due to A. simplex infection. Most cases of anisakiasis in Europe and Japan have been reported to be caused by Anisakis type I, particularly A. simplex [4]. However, anisakiasis due to A. pegreffii infection has also been reported from humans in Italy [50,51]. A. simplex might penetrate and migrate to fish muscle [52], which may explain the higher cases of anisakiasis due to A. simplex infection. Anisakiasis due to A. typica has not been reported. Umehara et al. [20] reported that A. typica so far has only received limited attention and is not widely recognized, thus its zoonotic impact has not been well documented. However, Palm et al. [25] reported that A. typica was not only found on the surface of gastrointestinal tract but it might also penetrate muscle of fish. In the present study, though not common, an Anisakis larva was observed to migrate into the musculature, indicating that the parasite has the potential to infect humans through consumption of uncooked food. In Indonesia, reports about anisakiasis in humans were suggested by Uga et al. [9] using a seroepidemiological approach of inhabitants in East Java and revealed that about 11% of samples who visited hospital showed positive results for Anisakis antibodies. The species of Anisakis spp. was not determined, but it might be possible that the parasite was A. typica since this species is widely distributed in tropical waters, compared to the well known causative agent of anisakiasis by A. simplex and A. pegreffii which have limited geographical distribution in cold waters. However, it might also be possible that Anisakis spp. could be from imported raw materials. Yoshinaga et al. [53] reported the presence of A. pegreffii in amberjack Seriola dumerili imported from China to Japan as mariculture seedlings.
In conclusion, based on PCR-RFLP and sequencing, all the Anisakis examined were A. typica, indicating the predominance of this species in the Southern Makassar Strait, Indonesia. Sequencing and phylogenetic tree analyses of Anisakis type I in ITS1-5.8S-ITS2 and mtDNA cox2 regions showed that the present samples were in the same cluster as A. typica published in GenBank. However, differences of 4 bp in nucleotides in ITS1 region and broad divergence consisting of 2 subgroups in the mtDNA cox2 of Anisakis from K. pelamis and A. thazard indicated the existence of A. typica sibling species in that area.
ACKNOWLEDGMENTS
This study was partially supported by Directorate General of Higher Education, Indonesia, through Indonesian National Strategic Research Grant (HIBAH STRANAS), grant no. 510/SP2H/PP/DP2M/VII/2010 (24 July 2010), and Academic Recharging Program conducted at The University of Tokyo, Japan. We would like to thank Prof. Tomoyoshi Yoshinaga, Assist. Prof. Hiroshi Yokoyama, and Dr. Daniel Grabner for comments and suggestions during molecular study at The Laboratory of Fish Diseases, The University of Tokyo, Japan.
Notes
There is no conflict of interest related with this study.

