Abstract
Cryptosporidium spp., ubiquitous enteric parasitic protozoa of vertebrates, recently emerged as an important cause of economic loss and zoonosis. The present study aimed to determine the distribution and species of Cryptosporidium in post-weaned and adult pigs in Shaanxi province, northwestern China. A total of 1,337 fresh fecal samples of post-weaned and adult pigs were collected by sterile disposable gloves from 8 areas of Shaanxi province. The samples were examined by Sheather’s sugar flotation technique and microscopy at×400 magnification for Cryptosporidium infection, and the species in positive samples was further identified by PCR amplification of the small subunit (SSU) rRNA gene. A total of 44 fecal samples were successfully amplified by the nested PCR of the partial SSU rRNA, with overall prevalence of 3.3%. The average prevalence of Cryptosporidium infection in each pig farms ranged from 0 to 14.4%. Species identification by sequencing of SSU rRNA gene revealed that 42 (3.1%) samples were Cryptosporidium suis and 2 (0.15%) were Cryptosporidium scrofarum. C. suis had the highest prevalence (7.5%) in growers and the lowest in breeding pigs (0.97%). C. suis was the predominant species in pre-weaned and adult pigs, while C. scrofarum infected pigs older than 3 months only. A season-related difference of C. suis was observed in this study, with the highest prevalence in autumn (5.5%) and the lowest (1.7%) in winter. The present study provided basic information for control of Cryptosporidium infection in pigs and assessment of zoonotic transmission of pigs in Shaanxi province, China.
-
Key words: Cryptosporidium, pig, SSU rRNA, Shaanxi province, China
Cryptosporidium, the worldwide distributed protozoans, have been recognized with 27 species and more than 70 genotypes that can infect humans and animals, including ruminants, amphibians, and birds, leading to serious enteritis even death in immunocompromised hosts and neonates [
1-
3]. Of susceptible animals, the pig is considered to be one of the main animal reservoir hosts of
Cryptosporidium [
4]. From pigs, 6
Cryptosporidium species have been isolated, namely,
C. suis,
C. parvum,
C. muris,
C. andersoni,
C. scrofarum (formerly named as
Cryptosporidium pig genotype II) [
5], and C.
tyzzeri (formerly named as
Cryptosporidium mouse genotype I) [
6]. Meanwhile, experimental infection studies demonstrated that pigs were also susceptible to infections with
C. hominis and
C. meleagridis, which suggested that pigs could be a source of infection for humans and other animals, posing an invisible threat to human security [
7-
9].
Cryptosporidium infection in pigs occurs mainly through an oral-fecal route. Despite the fact that it does not always cause clinical signs, severe diarrhea, weight loss, and even death can appear in neonatal and immunodeficient pigs [
10,
11]. Considering that previous studies around the world have indicated pigs as the main host for
C. suis and
C. scrofarum, and that humans are infected with
C. suis and
C. scrofarum, these 2 pig-adapted
Cryptosporidium species are potentially zoonotic [
12,
13]. The prevalence of
Cryptosporidium infection in pigs have been reported in several provinces in China, with the highest in Anhui (56.8%) and lowest in Henan (8.2%) provinces [
14,
15]. The
Cryptosporidium species have been identified in each research province, as mainly
C. suis and
C. scrofarum. Another species named
C. tyzzeri was found only in eastern China [
16]. However, it is unclear whether the pig naturally infected. Therefore, the present study aimed to determine the species and genotypes of
Cryptosporidium in domestic pigs in Shaanxi province, northwestern China by DNA sequencing of the SSU rRNA gene, and to elucidate the public health significance of pigs in this province.
From May 2011 to May 2014, a total of 1,337 fresh fecal samples of post-weaned and adult pigs (
Tables 1,
2) were collected from 9 intensive farms located in 8 counties/districts in Shaanxi province by sterile disposable gloves. Samples were firstly examined by Sheather’s sugar flotation technique and microscopy at×400 magnification. The microscopically positive samples were stored in 2.5% potassium dichromate. All microscopy-positive samples were then used to identify species/genotypes using the nested PCR and sequencing. Genomic DNA was extracted using the E.Z.N.A.
® Stool DNA Kit (OMEGA Biotek Inc., Doraville, Georgia, USA) according to the manufacturer’s instructions, and stored at -20˚C until further processed.
A 2-step nested PCR amplification of the small subunit (SSU) rRNA was used to determine species/genotypes of
Cryptosporidium. The procedure of Xiao et al. [
17] was followed except Taq polymerase-KOD FX Neo (Toyobo, Japan). Negative (without DNA template) and positive controls were included in each amplification running. Each secondary amplification was detected by 1.5% (w/v) agarose gel stained with ethidium bromide, and the positive amplicons were directly sequenced with secondary PCR primers on an ABI 3370 DNA sequencer at Sangon Company (Shanghai, China). Precise of the data was ensured by sequencing in both directions and hand assembly using DNAStar software as a sequence editor [
18]. Sequences from all samples (n=44) in this study were deposited in GenBank database under accession nos. KJ790201 to KJ790244.
Assembled sequences were identified into species/genotype in GenBank™ database using the BLAST (
http://www.ncbi.nlm.nih.gov) and phylogenetic analyses. The phylogenetic tree was reconstructed using the Neighbor-joining (NJ) method implemented in Mega 4.0 [
19] and the Kimura 2-parameter model. The consensus tree was obtained after bootstrap analysis, with 1,000 replications. The phylogenetic analysis based on SSU rRNA gene utilized
C. suis (GenBank accession no. JQ936485) and
C. scrofarum (no. JX424840) as ingroups, and
Plasmodium ovale (KF018658) as the outgroup. The prevalence differences of
Cryptosporidium oocysts among the factors of origins, cultivation modes, seasons, and ages were evaluated using Regression Analysis in Statistical Product and Service Solutions (SPSS) with 95% confidence intervals (CI). The probability level (P) of<0.05 was regarded as statistically significant.
Of 1,337 fresh fecal samples, 44 were successfully detected by both microscopy (
Fig. 1) and nested PCR, with an overall prevalence of 3.3%. The average prevalence of
Cryptosporidium infection in each pig farm ranged from 0 to14.4%. The farm in Wugong county using Fermentation Bed breeding conditions had the highest prevalence of 14.4% (27/187), whereas the average prevalence of the others under conventional breeding techniques was 1.5% (17/1,150). No infection was found in pigs from farms in Ankang and Xianyang counties. Statistical analysis showed the prevalence between different pig farms fluctuated greatly (
P<0.05). For different age groups of pigs, the prevalence were 1.3%, 1.4%, 7.5%, and 2.8% for breeding pigs (>6 months), fatteners (3-6 months), growers (1-2 months), and post-weaners (3 weeks-1 month), respectively. The difference of
Cryptosporidium infection rates between different age categories was statistically significant (
P<0.05). The prevalence of
Cryptosporidium infection in 4 seasons was significantly different (
P<0.05), with the highest in autumn (5.9%) and the lowest in winter (1.7%) (
Table 2).
The sequence analysis and phylogenetic analysis (
Fig. 2) based on SSU rRNA gene locus revealed the presence of 2
Cryptosporidium species, namely
C. suis (42/44) and
C. scrofarum (2/44).
C. suis was found in all
Cryptosporidium-positive areas, all seasons, and age groups, but
C. scrofarum was detected only in pigs older than 3 months in autumn in Wugong and Wuquan counties.
C. suis had the highest prevalence (7.5%) in growers and the lowest in breeding pigs (0.97%). A season-related difference of
C. suis was observed in this study, with the highest in autumn (5.5%) and the lowest (1.7%) in winter. Pairwise comparison of obtained sequences indicated that no variation was observed within isolates of
C. scrofarum, but 2 sequence types of
C. suis isolates were detected. For sequences of
C. suis isolates, only 1 Ins (G/-) was found in isolate CWQ4 from Wuquan.
The present study indicated
Cryptosporidium infection in pigs in Shaanxi province, with an overall infection rate of 3.3%, showing a lower infection rate than that in Henan (8.2%) and Anhui (56.8%) provinces, China [
20,
21]. Compared with the prevalence of
Cryptosporidium infection in pigs in other countries, it was relatively lower than that in western Australia (6.0%), northeastern Spain (22.5%), and Denmark (16%, 31%, and 100% for sows, piglets, and weaners) [
22-
26], but higher than that in Germany with the prevalence of 1.4% [
27]. Many factors have been considered to influence the prevalence of pig cryptosporidiosis among different areas and countries, including pig health status, age categories, breeding mode, and management style [
28,
29].
Cryptosporidium species have been molecularly genotyped in pigs in many countries, including China [
20,
22,
23,
30].
C. suis,
C. scrofarum,
C. muris,
C. tyzzeri,
C. parvum, and
C. andersoni were successfully isolated and identified [
25,
31-
35]. However, DNA sequencing of the SSU rRNA gene indicated only 2
Cryptosporidium species in this study, namely
C. suis and
C. scrofarum, and
C. suis was the predominant species in pre-weaned and adult pigs. These findings further confirmed that
C. suis and
C. scrofarum were the most common species in pigs [
12,
13,
30]. Although the 2 main species of human cryptosporidiosis, namely
C. parvum and
C. hominis, were not detected, both
C. suis and
C. scrofarum are also important zoonotic pathogens [
13,
36,
37]. In fact, the oocysts of 2 species have been found in water environment in some areas of China, including the source of drinking water companies, tap water as well as the wastewater nearby pig farms [
38-
40]. Therefore, pigs in Shaanxi province would pose an invisible threat to human security because of their asymptomatic infection and close contact with humans and water due to their huge numbers and economic importance. To further evaluate the public concern of pigs in this province, extensive molecular epidemiological surveys for better understanding of the transmission dynamics should be studied in the future.
Interestingly, season-related and age-related differences in
Cryptosporidium infection were observed in this study, with the highest prevalence in autumn (5.9%) and the lowest (1.7%) in winter.
C. suis predominantly infected younger piglets less than 3 months, while
C. scrofarum infected only pigs older than 3 months. Such season-specific and age-specific susceptibility were consistent with findings of some previous studies [
41,
42]. These results provided basic data for control of cryptosporidiosis in pigs in different seasons and age categories.
In conclusion, Cryptosporidium infection was observed in pigs in Shaanxi province, with the highest prevalence in autumn and the lowest in winter. C. suis and C. scrofarum were identified by sequencing of the SSU rRNA gene, with C. suis as the predominant species. The present study provided basic information for control of Cryptosporidium infection in pigs and assessment of zoonotic transmission source of pigs in Shaanxi province, China.
Notes
-
The authors declare that there is no conflict of interests in this paper.
This work was supported, in part, by the National Natural Science Foundation of China (grant no. 31101805), the Program for New Century Excellent Talents in University (NCET- 13-0489), the Fund for Basic Scientific Research (ZD2012010), and the Open Funds of th e State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SKLVEB2013KFKT007), China.
Fig. 1.
Cryptosporidium oocysts detected in fecal samples of pigs. Unstained.
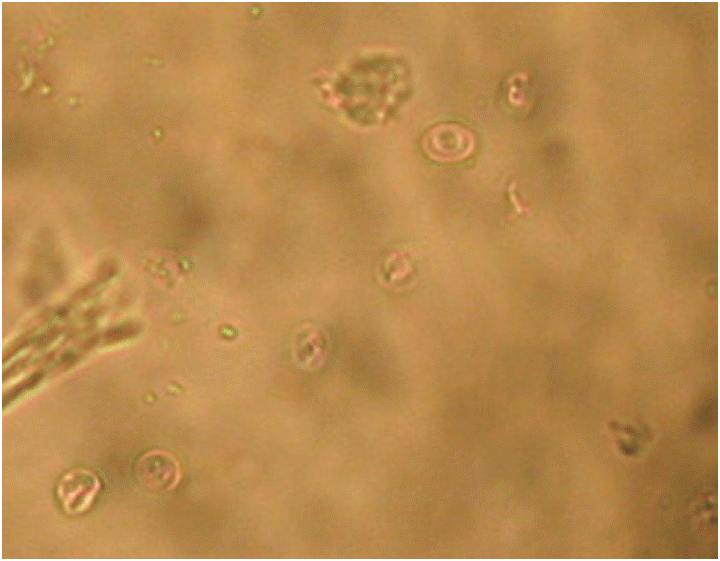
Fig. 2.Phylogenetic tree of representative samples reconstructed using the Neighbor-joining (NJ) method and the Kimura 2-parameter model. The consensus tree was obtained after bootstrap analysis, with 1,000 replications.
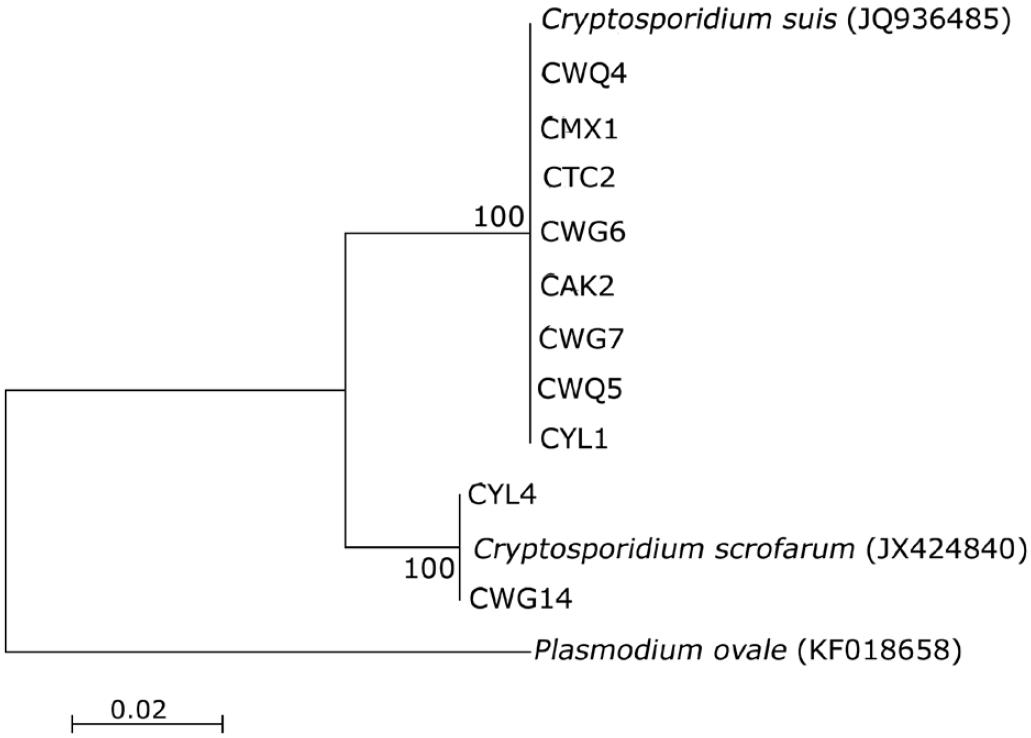
Table 1.Information of representative Cryptosporidium isolates in the present study
Table 1.
|
Sample code |
Location |
Age group |
GenBank accession no. |
|
CMX1 |
Meixian |
Grower |
KJ790236 |
|
CTC2 |
Tongchuan |
Grower |
KJ790244 |
|
CAK2 |
Ankang |
Fattener |
KJ790235 |
|
CWQ4 |
Wuquan |
Grower |
KJ790239 |
|
CWQ5 |
Wuquan |
Post-weaner |
KJ790234 |
|
CYL1 |
Yangling |
Grower |
KJ790242 |
|
CYL4 |
Yangling |
Breeding pig |
KJ790201 |
|
CWG6 |
Wugong |
Post-weaner |
KJ790243 |
|
CWG7 |
Wugong |
Post-weaner |
KJ790237 |
|
CWG14 |
Wugong |
Post-weaner |
KJ790202 |
Table 2.Prevalence and risk factors associated with Cryptosporidium infection in pigs in Shaanxi province, China
Table 2.
|
Factor |
Category |
No. examined |
No. positive (%) |
C. suis (%) |
C. scrofarum (%) |
|
Season |
|
|
|
|
|
|
Spring |
148 |
3 (2.0) |
3 (2.0) |
0 |
|
Summer |
404 |
9 (2.2) |
9 (2.2) |
0 |
|
Autumn |
438 |
26 (5.9) |
24 (5.5) |
2 (0.46) |
|
Winter |
347 |
6 (1.7) |
6 (1.7) |
0 |
|
Age |
|
|
|
|
|
|
3 weeks-1 month |
252 |
7 (2.8) |
7 (2.8) |
0 |
|
1-3 months |
358 |
27 (7.5) |
27 (7.5) |
0 |
|
3-6 months |
417 |
6 (1.4) |
5 (1.2) |
1 (0.24) |
|
> 6 months |
310 |
4 (1.3) |
3 (0.97) |
1 (0.32) |
|
Location |
|
|
|
|
|
|
Ankang |
76 |
0 |
0 |
0 |
|
Wugong |
187 |
27 (14.4) |
26 (13.9) |
1 (0.53) |
|
Wuquan |
185 |
5 (2.7) |
4 (2.2) |
1 (0.54) |
|
Yangling |
448 |
5 (1.1) |
5 (1.1) |
0 |
|
Xianyang |
92 |
0 |
0 |
0 |
|
Meixian |
121 |
3 (2.5) |
3 (2.5) |
0 |
|
Tongchuan |
124 |
3 (2.4) |
3 (2.4) |
0 |
|
Xi’an |
104 |
1 (0.96) |
1 (0.96) |
0 |
|
Total |
1,337 |
44 (3.3) |
42 (3.1) |
2 (0.15) |
References
- 1. Jex AR, Pangasa A, Campbell BE, Whipp M, Hogg G, Sinclair MI, Stevens M, Gasser RB. Classification of Cryptosporidium species from patients with sporadic cryptosporidiosis by use of sequence-based multilocus analysis following mutation scanning. J Clin Microbiol 2008;46:2252-2262.
- 2. Tomazic ML, Maidana J, Dominguez M, Uriarte EL, Galarza R, Garro C, Florin-Christensen M, Schnittger L. Molecular characterization of Cryptosporidium isolates from calves in Argentina. Vet Parasitol 2013;198:382-386.
- 3. Xiao L. Molecular epidemiology of Cryptosporidiosis: an update. Exp Parasitol 2010;124:80-89.
- 4. Leoni F, Amar C, Nichols G, Pedraza-Díaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 2006;55:703-707.
- 5. Kváč M, Kestřánová M, Pinková M, Kvĕtoñová D, Kalinová J, Wagnerová P, Kotková M, Vítovec J, Ditrich O, McEvoy J, Stenger B, Sak B. Cryptosporidium scrofarum n. sp. (Apicomplexa: Cryptosporidiidae) in domestic pigs (Sus scrofa). Vet Parasitol 2013;191:218-227.
- 6. Yui T, Nakajima T, Yamamoto N, Kon M, Abe N, Matsubayashi M, Shibahara T. Age-related detection and molecular characterization of Cryptosporidium suis and Cryptosporidium scrofarum in pre- and post-weaned piglets and adult pigs in Japan. Parasitol Res 2014;113:359-365.
- 7. Sheoran A, Wiffin A, Widmer G, Singh P, Tzipori S. Infection with Cryptosporidium hominis provides incomplete protection of the host against Cryptosporidium parvum. J Infect Dis 2012;205:1019-1023.
- 8. Akiyoshi DE, Dilo J, Pearson C, Chapman S, Tumwine J, Tzipori S. Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infect Immun 2003;71:1828-1832.
- 9. Darabus G, Olariu R. The homologous and interspecies transmission of Cryptosporidium parvum and Cryptosporidium meleagridis. Pol J Vet Sci 2003;6:225-228.
- 10. Bouzid M, Hunter PR, Chalmers RM, Tyler KM. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev 2013;26:115-134.
- 11. Omidian Z, Ebrahimzadeh E, Shahbazi P, Asghari Z, Shayan P. Application of recombinant Cryptosporidium parvum P23 for isolation and prevention. Parasitol Res 2014;113:229-237.
- 12. Kvác M, Kvetonová D, Sak B, Ditrich O. Cryptosporidium pig genotype II in immunocompetent man. Emerg Infect Dis 2009;15:982-983.
- 13. Xiao L, Bern C, Arrowood M, Sulaiman I, Zhou L, Kawai V, Vivar A, Lal AA, Gilman RH. Identification of the Cryptosporidium pig genotype in a human patient. J Infect Dis 2002;185:1846-1848.
- 14. Qiu SX, Lu QB, Qi M, Zhang XH, Duan ZX, Nin CS, Jian FC, Zhang LX. The prevalence of Cryptosporidium spp in pigs in Henan province. Chin J Zoonosis 2008;24:481-482. (in Chinese).
- 15. Zhao C, Li P. Epidemiologic survey of pig cryptosporidiosis in Fengtai county in Anhui province. Chin J Vet Parasitol 2003;11:42-44. (in Chinese).
- 16. Chen F, Huang K. Prevalence and phylogenetic analysis of Cryptosporidium in pigs in eastern China. Zoonoses Public Health 2007;54:393-400.
- 17. Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol 2001;67:1097-1101.
- 18. Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol 2000;132:71-91.
- 19. Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24:1596-1599.
- 20. Wang R, Qiu S, Jian F, Zhang S, Shen Y, Zhang L, Ning C, Cao J, Qi M, Xiao L. Prevalence and molecular identification of Cryptosporidium spp. Parasitol Res 2010;107:1489-1494.
- 21. Zhao CC, Li PY. Epidemiologic survey on pig cryptosporidiosis in Fengtai county in Anhui province. Chin J Vet Parasitol 2003;11:42-44. (in Chinese).
- 22. Ryan UM, Samarasinghe B, Read C, Buddle JR, Robertson ID, Thompson RC. Identification of a novel Cryptosporidium genotype in pigs. Appl Environ Microbiol 2003;69:3970-3974.
- 23. Maddox-Hyttel C, Langkjaer RB, Enemark HL, Vigre H. Cryptosporidium and Giardia in different age groups of Danish cattle and pigs-occurrence and management associated risk factors. Vet Parasitol 2006;141:48-59.
- 24. Suárez-Luengas L, Clavel A, QuÍlez J, GoÑi-Cepero MP, Torres E, Sánchez-Acedo C, del Cacho E. Molecular characterization of Cryptosporidium isolates from pigs in Zaragoza (northeastern Spain). Vet Parasitol 2007;148:231-235.
- 25. Zintl A, Neville D, Maguire D, Fanning S, Mulcahy G, Smith HV, De Waal T. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 2007;134:1575-1582.
- 26. Nguyen ST, Honma H, Geurden T, Ikarash M, Fukuda Y, Huynh VV, Nguyen DT, Nakai Y. Prevalence and risk factors associated with Cryptosporidium oocyst shedding in pigs in Central Vietnam. Res Vet Sci 2012;93:848-852.
- 27. Wieler LH, Ilieff A, Herbst W, Bauer C, Vieler E, Bauerfeind R, Failing K, Klös H, Wengert D, Baljer G, Zahner H. Prevalence of enteropathogens in suckling and weaned piglets with diarrhoea in southern Germany. J Vet Med B Infect Dis Vet Public Health 2001;48:151-159.
- 28. Roepstorff A, Nansen P. Epidemiology and control of helminth infections in pigs under intensive and non-intensive production systems. Vet Parasitol 1994;54:69-85.
- 29. Damriyasa IM, Bauer C. Prevalence and age-dependent occurrence of intestinal protozoan infections in suckling piglets. Berl Münch Tierärztl Wochenschr 2006;119:287-290.
- 30. Zhang W, Yang F, Liu A, Wang R, Zhang L, Shen Y, Cao J, Ling H. Prevalence and genetic characterizations of Cryptosporidium spp. in pre-weaned and post-weaned piglets in Heilongjiang Province, China. PLoS One 2013;8:e67564.
- 31. Morgan UM, Buddle JR, Armson A, Elliot A, Thompson RCA. Molecular and biological characterisation of Cryptosporidium in pigs. Australian Vet J 1999;77:44-47.
- 32. Guselle NJ, Appelbee AJ, Olson ME. Biology of Cryptosporidium parvum in pigs: from weaning to market. Vet Parasitol 2003;113:7-18.
- 33. Ryan UM, Monis P, Enemark HL, Sulaiman I, Samarasinghe B, Read C, Buddle R, Robertson I, Zhou L, Thompson RC, Xiao L. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J Parasitol 2004;90:769-773.
- 34. Langkjaer RB, Vigre H, Enemark HL, Maddox-Hyttel C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology 2007;134:339-350.
- 35. Hsu BM, Wun HY, Hsu CL. Detection and species identification of Cryptosporidium from Taiwan feeding animals. J Parasitol 2008;94:252-256.
- 36. Leoni F, Amar C, Nichols G, Pedraza-Díaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. 2006, 55:pp 703-707.
- 37. Kvác M, Hanzlíková D, Sak B, Kvetonová D. Prevalence and age-related infection of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet Parasitol 2009;160:319-322.
- 38. Xiao S, An W, Chen Z, Zhang D, Yu J, Yang M. Occurrences and genotypes of Cryptosporidium oocysts in river network of southern-eastern China. Parasitol Res 2012;110:1701-1709.
- 39. Feng Y, Li N, Duan L, Xiao L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol 2009;47:153-157.
- 40. Feng Y, Zhao X, Chen J, Jin W, Zhou X, Li N, Wang L, Xiao L. Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in Shanghai, China. Appl Environ Microbiol 2011;77:3609-3616.
- 41. Jeníková M, Nĕmejc K, Sak B, Kvĕtoñová D, Kváč M. New view on the age-specificity of pig Cryptosporidium by species-specific primers for distinguishing Cryptosporidium suis and Cryptosporidium pig genotype II. Vet Parasitol 2011;176:120-125.
- 42. Yoshiuchi R, Matsubayashi M, Kimata I, Furuya M, Tani H, Sasai K. Survey and molecular characterization of Cryptosporidium and Giardia spp. in owned companion animal, dogs and cats, in Japan. Vet Parasitol 2010;174:313-316.


