Cloning and Iron Transportation of Nucleotide Binding Domain of Cryptosporidium andersoni ATP-Binding Cassette (CaABC) Gene
Article information
Abstract
Cryptosporidium andersoni ATP-binding cassette (CaABC) is an important membrane protein involved in substrate transport across the membrane. In this research, the nucleotide binding domain (NBD) of CaABC gene was amplified by PCR, and the eukaryotic expression vector of pEGFP-C1-CaNBD was reconstructed. Then, the recombinant plasmid of pEGFP-C1-CaNBD was transformed into the mouse intestinal epithelial cells (IECs) to study the iron transportation function of CaABC. The results indicated that NBD region of CaABC gene can significantly elevate the transport efficiency of Ca2+, Mg2+, K+, and HCO3- in IECs (P<0.05). The significance of this study is to find the ATPase inhibitors for NBD region of CaABC gene and to inhibit ATP binding and nutrient transport of CaABC transporter. Thus, C. andersoni will be killed by inhibition of nutrient uptake. This will open up a new way for treatment of cryptosporidiosis.
Cryptosporidiosis is a zoonotic parasitic disease caused by Cryptosporidium infection [1]. Cryptosporidium usually parasitizes the epithelial cells of the gastrointestinal tract in the host, and causes severe diarrhea. So far, Cryptosporidium animal models have been established [2], and the treatment of cryptosporidiosis has been studied in vitro [3-6]. However, there is still no effective drug for treating cryptosporidiosis. A possible reason is that Cryptosporidium have multidrug ATP-binding cassette (ABC) transporters to inhibit anti-protozoal drugs and to enter the protozoa bodies [7,8]. It was also reported that ABC transporters use the energy of ATP binding and hydrolysis to drive the transport of various substrates across the cell membrane [3-5]. C. parvum ABC1 (CpABC1), C. parvum ABC2 (CpABC2), and C. parvum ABC3 (CpABC3) which have been proved in C. parvum can transport different substrates across the cells through the energy of ATP binding and hydrolysis [9-12].
Cryptosporidium andersoni usually parasitizes the ruminants and people [13]. So far, CaABC for the transportation of nutrients and the characteristics of resistant drugs were not reported. Two transmembrane domains (TMDs) and 2 nucleotide-binding domains (NBDs) constitute the basic architecture of ABC transporters, and TMDs can provide the substrate translocation pathway across the cell membrane, while the NBDs bind and hydrolyze ATP to provide energy for active transport [14,15]. Walker et al. [16] showed that NBD region is in the conservative domain of ABC protein in C. parvum. However, there is no research of the existence of NBD region in CaABC protein and its exact function. In the study, our first objective was to amplify the NBD region of CaABC gene and to construct the CaNBD eukaryotic recombinant plasmid. Our second objective was to establish a cell model carrying CaNBD and to study the role of CaNBD. These results will provide the basis for nutrient transport of CaABC transporter in the presence of ATPase inhibitor.
All of experimental procedures on animals were in accordance with the recommendations of the guidelines of the Chinese Association for Laboratory Animal Science. The feces of cows infected with C. andersoni were obtained from a cattle farm in Hefei. C. andersoni oocysts were separated and washed 3 times with PBS, and shocked in a vortex mixer 30 min after adding 500 μl oocyst lysate, repeatedly, then freeze-thawed in -70˚C 3 times. Genomic DNA of C. andersoni was extracted with the DNA extraction kit (Omega, New York, USA) according to the instructions of the manufacturer. The primer of NBD region of CaABC gene was designed according to Perkins [7]. The primer with promoter ATG, terminator TAA, and enzyme cut sites BglII and EcoRI (TaKaRa, Dalian, China) was synthesized (Sangon Biotech, Shanghai, China). The forward primer was 5´GAAGATCTATGGTAGGTGAAACTGGTAGTGGTAAATCTAC, the reverse primer was 5´CGGAATTCTTAATCTAGAGAAGATGTAGCTTCATC.
Cloning and identification of NBD region of CaABC gene were performed by PCR. The product was examined using 1.0% agarose gel electrophoresis and observed with the gel imaging system (BIO-RAD, Hercules, California, USA) and extracted with gel extraction kit (Sangon Biotech, Shanghai, China) according to the instructions of the manufacturer. The product was linked to a pMD19-T clone vector (TaKaRa, Dalian, China), and transformed into E. coli DH5α (Sangon Biotech, Shanghai, China). The plasmid of positive colony was extracted by PCR and was sequenced. The constructed clone vector was named as pMD19-T- CaNBD.
The sequencing CaNBD and eukaryotic expression vector pEGFP-C1 were digested by restriction double-enzyme EcoRI and BglII. The products were analyzed by electrophoresis and respectively extracted with gel extraction kit according to the instructions of manufacturer. The target gene was linked to linear pEGFP-C1 vector under the action of T4 DNA ligase (TaKaRa, Dalian, China). After transfection, PCR and sequence identification, the pEGFP-C1-CaNBD recombinant plasmid was obtained.
In transfection experiment, the experiment was divided into 3 groups: transfection group is the IECs that transfected a recombinant plasmid pEGFP-C1-CaNBD; the control group is the IECs that transfected empty plasmid pEGFP-C1; the blank group is IEC cells without plasmid. Before transfection at 24 hr, primary IECs were cultured at 37˚C, 5% CO2 for 18-48 hr in a 24-well plate (1.5×105 cells/well) which was coated by 1% gelatin. A density of 80-90% IECs was transfected. Then, the cells were analyzed with the fluorescence microscope (Olympus-CK40, Tokyo, Japan). The medium in 3 groups were collected to analyze the extracellular ions concentration. The cells in 3 groups were digested with 0.25% trypsin, collected, and broken up by ultrasonic cell disruptor (VCX-500, Sonics, Danbury, Connecticut, USA). Intracellular and extracellular ion (Ca2+, Mg2+, K+, and HCO3-) concentrations were analyzed with calcium, magnesium, potassium assay kit, and HCO3- reagent kit (Jiancheng Inc., Nanjing, China) according to the instructions of manufacturer by automatic biochemistry analyzer (Hitachi-7060, Tokyo, Japan). The experiments were carried out 3 times. Statistical analyses were conducted using the SPSS version 17 software (Version 17.0; SPSS, Inc., Shanghai, China). All the values were considered significant at P<0.05.
NBD region of CaABC gene was amplified by PCR. A DNA band about 427 bp was observed, which was in accordance with the expected result (Fig. 1). It was clear that NBD region of CaABC gene was successfully amplified. In order to identify NBD region of CaABC protein, the product was sequenced and analyzed. The results showed that the purpose fragment was 433 bp (Fig. 2). The restriction enzyme sites, the promoter and terminator were 22 bp. The fragment of NBD region of CaABC gene was 411 bp; it was more than 6 bp in comparison with Plasmodium glycoprotein (Pgp1) gene sequence.

Electrophoregram of PCR product of NBD region of CaABC gene. M: DNA marker; 1: a DNA band of NBD region of CaABC gene.
Nucleotide sequences of NBD region of CaABC gene were translated into a protein with 137 amino acids: VGETGSGKSTILKLLERIYKPQNGEIEYFGVTGGLLSDANIRELFAYVPQDCA
LFEGSIRENIVFGKLNASMNEIEEAAKRSAVNDFIESLPEKYDMAVGERGSRLSGGQRQRIAIARALIKGAPIVLLDEATSSLD.
Amino acid sequence of NBD region of CaABC gene was compared with the Pgp1 and C. parvum multidrug resistance-associated protein (Cp-MRP) by BLAST, respectively. The results showed that 9 amino acids of the Walker A motif in NBD region of CaABC gene in N-terminal amino acid sequence and 10 amino acids of Walker B in C-terminal amino acids were the same as Pgp1 (Fig. 3A) and Cp-MRP (Fig. 3B), and a NBD region of CaABC protein family exist, which is composed of 9 amino acids. Therefore, 411 bp nucleotides can be determined as ATP binding region sequence of CaABC protein, named as CaNBD.

Amino acid sequences of CaNBD. (A) CaNBD amino acid sequences compared with Pgp1. (B) CaNBD amino acid sequences compared with Cp-ATP.
Two DNA bands, about 4.7 kb and 433 bp, were observed in the study (Fig. 4). The former was the linear plasmid pEGFP-C1, and the latter was a purpose gene, which was in accordance with the expected result. The results proved that the recombinant eukaryotic plasmid pEGFP-C1-CaNBD was successfully constructed.
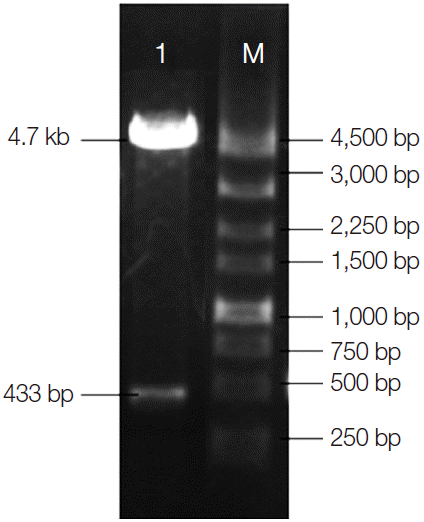
Electrophoregram of double digestion of pEGFP-C1-CaNBD. M: DNA marker; 1: a DNA band of pEGFP-C1-CaNBD double digestion.
Mouse IECs with enhanced green fluorescent protein (EGFP) gene in the transfection group (Fig. 5C) and control group (Fig. 5B) were observed after being transfected. Mouse IECs with EGFP gene were more in the control group (Fig. 5B) compared to the transfection group (Fig. 5C). Mouse IECs in blank group (Fig. 5A) was no green fluorescence; however, auto-fluorescence in a small amount of apoptosis IECs was observed. The results showed that green fluorescence was emitted by the EGFP gene from the introduced plasmid. The eukaryotic expression vector pEGFP-C1-CaNBD was successfully transfected into mouse IECs.
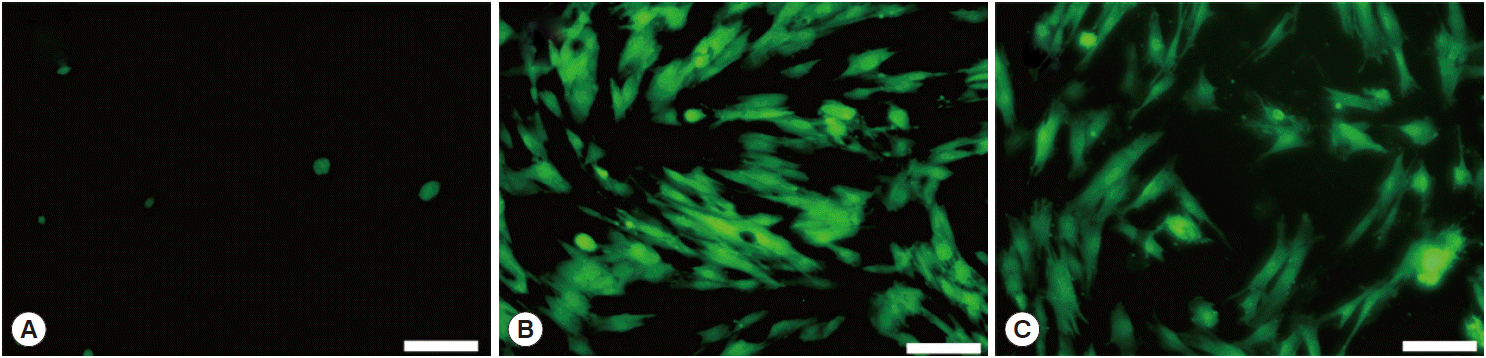
Expression of EGFP in different IECs groups. (A) The blank group (IECs without plasmid). (B) The control group (IECs transfected with empty plasmid pEGFP-C1). (C) Transfection group (IECs transfected with a recombinant plasmid pEGFP-C1-CaNBD). Bar=100 μm.
The intracellular Ca2+, Mg2+, K+, and HCO3- concentrations in the transfection group were significantly elevated in comparison with the black group and control group (P <0.05) (Fig. 6A); however, no significant difference was observed between the black group and control group (P >0.05). A significantly lower level of the extracellular Ca2+, Mg2+, K+, and HCO3- concentrations were present in the transfection group (P <0.05) in comparison with the black group and control group (Fig. 6B). Furthermore, the extracellular Ca2+, Mg2+, K+, and HCO3- concentrations in the control group were not significantly lower than in the black group (P >0.05).
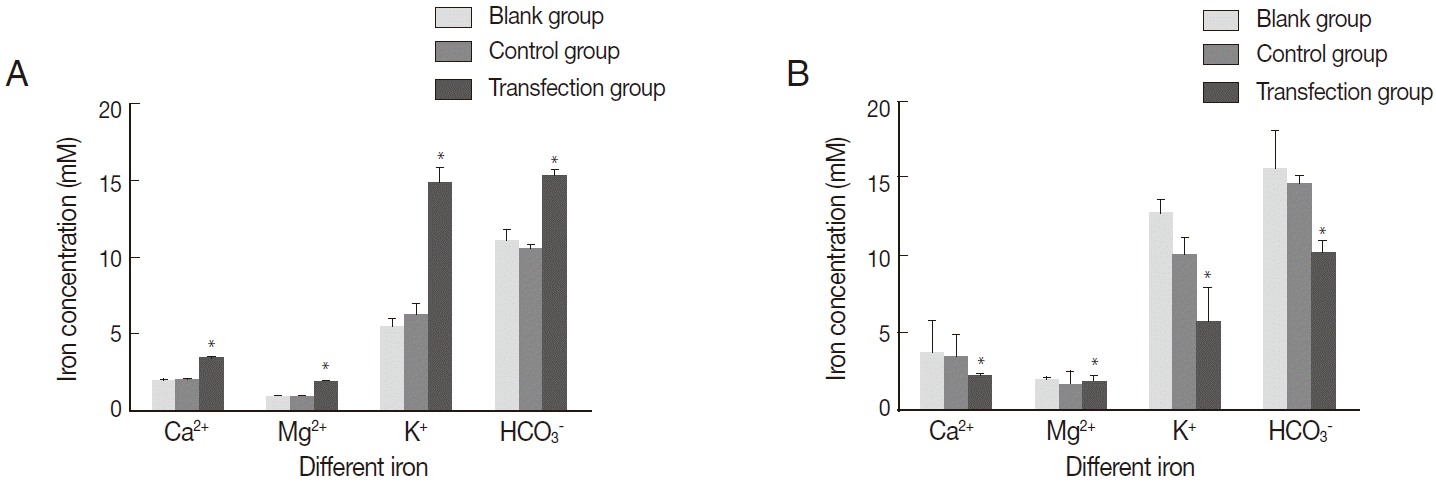
Change of intracellular and extracellular ions concentration. (A) Change of intracellular ions concentration. (B) Change of extracellular ions concentration. *P<0.05. Error bars represent SEM of 9 repeats.
The ABC proteins generally consisted of 4 regional; 2 hydrophobic transmembrane binding regions and 2 NBDs [14,15]. Walker et al. [16] clarified that the NBDs existed in the conservative area of ABC transporters, which contained 2 main motifs (Walker A and Walker B) and ABC protein characteristic motifs. In this study, 9 amino acids of the Walker A motif in NBD region of CaABC gene in N-terminal amino acid sequence and 10 amino acids of Walker B in C-terminal amino acids were the same as Pgp1 and Cp-MRP. The results were in agreement with previous studies. Therefore, the amplification of 411 bp gene is NBD region of CaABC gene.
CpABC protein is transport proteins located in fold structure feeding device membrane of Cryptosporidium, which has greatly relation with Cryptosporidium nutrient intake and waste drainage [17]. In this study, the recombinant plasmid pEGFP-C1-CaNBD was constructed, and imported into mouse IECs. Recombinant plasmid pEGFP-C1-CaNBD was successfully imported and expressed in mouse IECs by analyzing the results of recombinant plasmid sequencing and observing IECs fluorescence. The results showed that NBD region of CaABC gene could express validly in mouse IECs, and the ABC protein of Cryptosporidium research continue to expand through the cell model.
This study also showed the changes of ion concentration in IECs after NBD domain transformation. The mechanism may be that NBD region of CaABC transporter is responsible for ATP binding and hydrolysis and regulate gated substrate channels. Thus, NBD domain transformation in IECs was used for more ATP binding; gated substrates channels (including ion channel) were widely opened. Hence, the ion concentration in IECs by NBD domain transformation changed. The amplification and expression of NBD region of CaABC gene will provide an important basis for ABC protein gene complete sequence amplification and study of nutrient transport and multidrug resistance in IECs. It is expected to find the inhibitor to inhibit ATP binding to NBD region and transport processes with substrates. The eventual purpose is used for drug development and treatment of cryptosporidiosis.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31001019) and the Academic Backbone Training Project of Anhui Agricultural University (No. 2014XKPY-21). The authors sincerely thank Tao Sun and Wei Liu for the assistance during the preparation of the study.
Notes
The authors report no conflicts of interest with this study.
