Anisakis pegreffii Larvae in Sea Eels (Astroconger myriaster) from the South Sea, Republic of Korea
Article information
Abstract
Anisakis simplex sensu stricto (s.s.), Anisakis pegreffii, Anisakis berlandi (=A. simplex sp. C), and Anisakis typica are the 4 major species of Anisakis type I larvae. In the Republic of Korea (Korea), A. pegreffii, A. berlandi, and A. typica larvae in fish hosts has seldom been documented. In this study, molecular analysis was performed on Anisakis larvae from the sea eels (Astroconger myriaster), the major source of human anisakiasis in Korea, collected from Tongyeong City, a southern coastal area of Korea. All 20 sea eels examined were infected with Anisakis type I larvae (160 larvae; 8 per fish). Their species were analyzed using PCR-RFLP patterns and nucleotide sequences of internal transcribed spacers (ITS1, 5.8 subunit gene, and ITS2) and mitochondrial cytochrome c oxidase 2 (cox2). Most (86.8%; 112/129) of the Anisakis type I larvae were A. pegreffii, and 7.8% (10/129) were A. typica. The remaining 5.4% (7/129) was not identified. Thus, A. pegreffii is the major species of anisakid larvae in sea eels of the southern coast of Korea.
Human anisakiasis is caused mainly by the larvae of the genera Anisakis and PseudoTerranova [1]. The 2 most important species are Anisakis simplex sensu stricto (s.s.) and PseudoTerranova decipiens [1,2]. However, since 1999, a few human infections with Anisakis pegreffii larvae (a sibling species of A. simplex s.s.) have been reported by molecular analysis in Italy [3-6] and Japan [7,8]. Subsequently, human infections with A. pegreffii larvae were confirmed in 2015 in 15 of 16 clinical patients in the Republic of Korea (Korea), from whom the larvae were extracted by gastroduodenoscopy [9]. The number of human A. pegreffii cases may be much higher than hitherto considered.
The sources of human anisakiasis are marine fish and squids [1,2]. Many studies have reported on the taxonomy and classification of anisakid larvae recovered in these hosts. Several morphological types were described, including Anisakis type I, Anisakis type II, Terranova type A, Terranova type B, Contracaecum type A, and Contracaecum type B [1,10]. However, since the development of molecular techniques, it became apparent that even within a type several species or strains may be included. For example, Anisakis type I larvae include at least 4 different species: A. simplex s.s., A. pegreffii, and Anisakis berlandi (A. simplex sp. C), and Anisakis typica [11].
In Korea, studies have been performed on anisakid larvae in various marine fish species [12-19] including the sea eel (Astroconger myriaster), which is one of the most important sources of human anisakiasis in Korea [1]. In the sea eel, most studies reported the presence of Anisakis type I larvae (some authors interpreted them as A. simplex s.s. larvae), and the presence of A. pegreffii larvae has seldom been documented with the exception of a recent report that detected A. pegreffii larvae by molecular analysis of the gene [20]. In other fish species and squids, only 2 reports [18,19] described the presence of A. pegreffii larvae in fish and squids caught in Korea. Thus, the present study aimed to investigate the status of A. pegreffii larvae infection in the sea eels caught from the South Sea, Korea.
Twenty sea eels (A. myriaster) were purchased in March 2013 at Guri Agricultural and Marine Products Market, Gyeonggi-do, Korea. The sea eels were stated to have been caught near Tongyeong city in the south coast of Korea. In total, 160 Anisakis type I third-stage larvae (L3) were collected; 129 of these 160 larvae were molecularly analyzed.
The total genomic DNA (gDNA) was extracted by DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Identification to the species level was carried out by PCR-RFLP and sequencing of a 274-900 bp fragment of internal transcribed spacer (ITS1, 5.8 subunit rRNA, and ITS2) gene and a 582-629 bp fragment of the mitochondrial cytochrome c oxidase 2 (cox2) gene. A region of the nuclear ribosomal DNA (rDNA) was amplified using the ITS A (5'-GTC GAA TTC GTA GGT GAA CCT GCG GAA GGA TCA-3') and B (5'-GCC GGA TCC GAA TCC TGG TTA GTT TCT TTT CCT-3') (21) with Smart 2× PCR premix Taq (Solgent, Daejeon, Korea), containing 10 pmol of each primer and 30 ng of the total DNA. The mixture undergone initial denaturation at 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 1.15 min with a final extension at 72°C for 7 min. The cox2 gene was amplified using the primers 211F 5'-TTT TCT AGT TAT ATA GAT TGR TTY AT-3' and 210R 5'-CAC CAA CTC TTA AAA TTA TC-3' (22). The mixture was denatured at 94°C for 3 min, followed by 34 cycles at 94°C for 30 sec, 46°C for 1 min, and 72°C for 1.5 min, followed by post-amplification at 72°C for 10 min. The ITS PCR products (17 μl) were digested with 10 units of restriction endonuclease Hinf1 (1 µl) (Enzynomics, Daejeon, Korea) and 10× EZ-one buffer (2 µl) (Enzynomics) in a final volume of 20 µl at 37°C for 1 hr. The digested products were then separated by electrophoresis on 3% agarose gels containing 1 µg/ml ethidium bromide and visualized under ultraviolet light. The nucleotide sequences of ITS and cox2 regions were obtained from 3-6 larval specimens and aligned using the program Geneious v.6.0.3.
All of the sea eels examined were found to be infected with Anisakis larvae (20/20, 100%), and a total of 160 Anisakis type I larvae (8.0 per fish) were collected. The larvae had a boring tooth, a ventriculus, and a mucron. The anterior portion showed a prominent boring tooth in the cephalic region. In the digestive tract, the ventriculus level showed the simply connected esophagus, ventriculus, and intestine. The posterior portion showed a mucron in the caudal region.
Out of the 129 Anisakis larvae identified by PCR-RFLP analysis, an approximately 900 bp fragment was produced after amplification of the rDNA region (ITS1, 5.8 subunit, and ITS2). The PCR products were processed to identify the species with restriction enzyme Hinf1. Hinf1 is the most appropriate and the best known enzyme for anisakid larvae identification [7,23]. RFLP produced 2 patterns. One revealed 3 different fragments approximately between 250 and 500 bp (A. pegreffii) and the other displayed 2 different fragments between 250 bp and 1,000 bp (A. typica). Most (86.8%; 112/129) larvae were diagnosed as A. pegreffii, whereas 7.8% (10/129) were diagnosed as A. typica. The remaining 5.4% (7/129) could not be diagnosed (Fig. 1).

(A) PCR-RFLP patterns of the rDNA region spanning the ITS-1, 5.8S, and ITS-2, for A. pegreffii and A. typica from the sea eels. M, 100-bp ladder. Lanes 1 and 3, gDNA from Anisakis spp.; lane 2, A. pegreffii (370, 300, and 250 bp); lane 4, A. typica (350 and 620 bp). (B) The infection status of the sea eels with different species of Anisakis larvae (n=129).
The final confirmation of the species was made after sequencing of the ITS and cox2. Three randomly selected Anisakis type I larvae were undergone sequencing of the ITS region (274 bp), and all of the samples revealed 100% identity with A. pegreffii (GenBank no. AB277823) and 99.2% identity with A. simplex s.s. (no. AB277822) (Table 1). In addition, 6 randomly selected samples underwent cox2 sequencing of the 582-629 bp region. Results indicated that the larvae were identical (97.8-100%) with A. pegreffii (no. EU413958) rather than with A. simplex s.s. (AB517560) (94.9-95.5%) (Table 1). Based on this information, phylogenetic trees of anisakid nematodes based on ITS and cox2 sequences were constructed using the neighbor-joining method (Free Software Foundation, Boston, Massachusetts, USA) (Fig. 2).
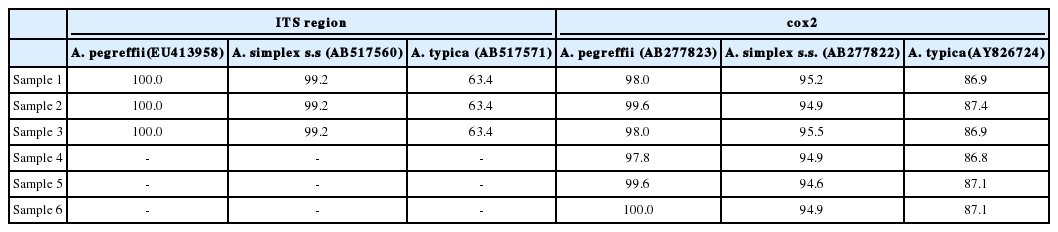
Nucleotide identity (%) of our specimens with the known Anisakis pegreffii, A. simplex s.s. and A. typica sequences in GenBank for the ITS region and cox2

Neighbor-Joining trees of Anisakis spp. based on (A) ITS region (274 bp) and (B) cox2 gene (511 bp) exploring the relationships among Anisakis spp. available in GenBank and Anisakis larvae collected from the sea eels in Korea.
Human anisakiasis is an important fish-borne parasitic zoonosis. The infection is caused by the ingestion of raw or improperly cooked fish infected by the larvae [1,2]. The overall infection rate of Anisakis larvae in the sea eels was 100%, with the infection density of 8 larvae per fish. As seen from its infection status, the sea eel is suspected as one of the most important paratenic fish hosts that may be related with human anisakiasis in Korea [1].
A. pegreffii was first reported from a Mediterranean monk seal in 1955 [24]. A. pegreffii was subsequently repeatedly reported in marine fish, squids, but rarely humans, in various localities including Europe, Australia, America, and Asia (including Japan, Chia, and Korea) [7,9,25]. Because the morphology of A. pegreffii larvae is almost indistinguishable from A. simplex s.s. larvae, electrophoretic analysis was first used for the differentiation [26]. Later, other molecular techniques, including PCR-RFLP and nucleotide sequencing, were developed, and these techniques are most popularly used [3-9].
Human A. pegreffii infection was first reported in Italy using PCR-RFLP [3]. The second was from Japan, in which 1 of 85 anisakid larvae extracted from human anisakiasis patients in Kyushu was identified as A. pegreffii by PCR-RFLP of the ITS region [7]. Thereafter, 11 human cases have been described in Italy [4-6]. Recently, 15 human infections with A. pegreffii were confirmed in Korea by nucleotide sequencing of the ITS region [9]. Therefore, at present, 28 molecularly confirmed human A. pegreffii infections have been reported in the literature.
With regard to the Anisakis larvae from fish in Korea, most previous reports designated the larvae simply as Anisakis type I [12-15], and sometimes A. simplex larvae (this means A. simplex complex) were used to designate them [16,17]. According to a recent report, the prevalence of Anisakis larvae was high; 52.3% (45 of 86) of fish from the East Sea, 76.6% (131 of 171) of fish from the South Sea, and 40.2% (37 of 92) of fish from the Yellow Sea [17]. In Busan, a port city on the South Sea, 2,537 fish and cephalopods were collected, and the overall infection rate of A. simplex larvae (i.e., A. simplex complex) was 34.3%, and the intensity was 17.1 and 5.9 larvae per fish and cephalopod, respectively, with the highest abundance in the spring season [16].
The diagnosis of A. pegreffii for the larvae from fish in Korea was first given to those collected from marine fish (Scomber japonicus and Trichiurus lepturus) and squids (Tadarodes pacificus) purchased from wholesale and retail markets in 2009 based on PCR-RFLP patterns of the ITS region [18]. It is noteworthy that most of the larvae were A. pegreffii (47/60 larvae; 78.3%) and A. typica (10/60; 16.7%), while only 1 (from a squid) of 60 larvae was A. simplex s.s. [18]. In contrast, a study of Anisakis larvae from the chum salmon (Oncorhynchus keta) caught in an eastern coastal area of Korea revealed that all 48 larvae analyzed by PCR-RFLP of mitochondrial DNA cox2 region were consistent with A. simplex s.s. [19]. Another study using PCR-RFLP and sequencing of ITS and mitochondrial cox1 regions reported that most of the larvae collected from 10 fish species from the Yellow Sea, South Sea, and East Sea were A. pegreffii, whereas very few were A. simplex s.s. [20]. Our study is consistent with 2 recent studies in Korea [18,20]. The significance of detecting A. typica larvae in sea eels remains to be further studied.
The geographical distribution of A. pegreffii larvae is Korea and Japan is interesting. In Japan, A. pegreffii larvae were more frequently detected than A. simplex s.s. in the western part (the East and South Seas of Korea), whereas A. simplex s.s. larvae dominated in the eastern part (the Pacific Ocean) [27]. In Korea, most larvae were A. pegreffii in various kinds of marine fish and squids [18,20]. In the present study, 86.8% (112/129) of the larvae were A. pegreffii and 7.8% (10/129 larvae) were A. typica. The predominance of A. pegreffii in the sea eels and other marine fish and cephalopods seems to be closely related to the predominance of human A. pegreffii infection (15 of 16 human cases) in Korea [9]. Molecular studies on the larvae extracted from human infections are needed in Korea.
Notes
We have no conflict of interest related to this work.