Allelic Diversity of MSP1 Gene in Plasmodium falciparum from Rural and Urban Areas of Gabon
Article information
Abstract
The present study determined and compared the genetic diversity of Plasmodium falciparum strains infecting children living in 2 areas from Gabon with different malaria endemicity. Blood samples were collected from febrile children from 2008 to 2009 in 2 health centres from rural (Oyem) and urban (Owendo) areas. Genetic diversity was determined in P. falciparum isolates by analyzing the merozoite surface protein-1 (msp1) gene polymorphism using nested-PCR. Overall, 168 children with mild falciparum malaria were included. K1, Ro33, and Mad20 alleles were found in 110 (65.5%), 94 (55.9%), and 35 (20.8%) isolates, respectively, without difference according to the site (P>0.05). Allelic families’ frequencies were comparable between children less than 5 years old from the 2 sites; while among the older children the proportions of Ro33 and Mad20 alleles were 1.7 to 2.0 fold higher at Oyem. Thirty-three different alleles were detected, 16 (48.5%) were common to both sites, and 10 out of the 17 specific alleles were found at Oyem. Furthermore, multiple infection carriers were frequent at Oyem (57.7% vs 42.2% at Owendo; P=0.04) where the complexity of infection was of 1.88 (±0.95) higher compared to that found at Owendo (1.55±0.75). Extended genetic diversity of P. falciparum strains infecting Gabonese symptomatic children and high multiplicity of infections were observed in rural area. Alleles common to the 2 sites were frequent; the site-specific alleles predominated in the rural area. Such distribution of the alleles should be taken into accounts when designing MSP1 or MSP2 malaria vaccine.
INTRODUCTION
Malaria, a disease mostly caused by Plasmodium falciparum, remains a major public health problem in sub-saharan Africa; where vulnerable groups are children and pregnant women [1]. The genetic complexity of P. falciparum and its ability to generate variants makes it a successful pathogen. Molecular markers, such as merozoite surface protein-1 (msp1) gene, reveal a spectrum of population structures in P. falciparum. Paul et al. [2] have hypothesized that genetic structure of P. falciparum is a function of transmission intensity in a given endemic area, but for other authors, this relationship may be non-linear [3]. Indeed, a positive non-linear association between low or intermediate altitude (in high malaria transmission areas) and an increase in P. falciparum msp1 gene block 2 polymorphisms generated by insertion/deletion of repeats was found [4]. The variable genetic structure of the pathogen has also been related to the development of antimalarial immunity in endemic areas. This variability significantly contributes to the parasite evasion of the immune response leading to the selection or dominance of certain genotypes in geographical different areas [5]. In another hand, individuals are often simultaneously infected by multiple parasite clones that are related to the transmission intensity, and were described as a factor determining the host immune status [5-8]. Analysis of P. falciparum clonal composition and its relationship with the transmission intensity in endemic areas is crucial to understand the dynamics of host-parasite interaction and the development of anti-malarial immunity.
In Gabon, since the deployment of ACTs, ITNs use, and IPTp-SP among pregnant women, a decline of malaria prevalence has been observed in different urban areas from 2000 to 2010 [9,10]. The spatial distribution of malaria in Gabon is heterogeneous, and its transmission intensity is probably governed by genetically distinct P. falciparum isolates which are suspected to be more virulent in some areas, as entomological data do not suggest any significant distribution in infected mosquito densities [10,11]. Few studies on the genetic diversity of Plasmodium isolates from Gabon are available, and comparisons according to malaria endemicity are lacking [12,13]. Furthermore, since the introduction of the new strategies in the country, limited information is available on the genetic diversity of circulating P. falciparum strains. Thus, the present study examined and compared the genetic diversity of P. falciparum isolates from children living in 1 urban and 1 rural areas of Gabon using a highly polymorphic gene encoding MSP1.
MATERIALS AND METHODS
Study areas
The study was conducted in Gabon where malaria transmission is perennial and predominantly due to P. falciparum. From February 2008 to January 2009, recruitment was carried out at the Centre Hospitalier Regional d’Oyem (CHRO) and the Centre de Santé Communautaire d’Owendo (CSCO) 25 km south of Libreville, the capital city of Gabon [14]. Malaria prevalence among febrile children was 39% and 11% at Oyem and Owendo, respectively, at the time of the sample collection [14].
Sample collection
This study was a part of a health facility-based project initiated by the Malaria National Control Program (MNCP), which aimed to evaluate malaria prevalence and to perform molecular analysis of P. falciparum isolates. Throughout the study, children under 11 years old, presenting at hospital for consultation with fever (axillary temperature ≥37.5˚C) or a history of fever in the preceding 48 hr, were enrolled [14]. Malaria diagnosis was performed by microscopy using the Lambaréné method, as previously described [15]. After parents or guardians have given consent, the blood samples of P. falciparum infected children (50 µl) were collected onto a filter paper for DNA extraction. For each patient, body temperature, history of fever, and sex were recorded. A single blood sample was taken for malaria diagnosis.
MSP1 gene analysis
DNA was extracted from the filter paper using the methanol extraction method [16]. The block 2 of msp1 gene was amplified by primary PCR followed by nested rounds of PCR using allele specific primers. Ro33, K1, and Mad20 alleles were determined. The PCR cycles and the primer sequences used are described elsewhere [17]. Allele-specific positive controls and DNA-free negative controls were included in each set of reactions. The amplicons were then run on 2.0% agarose gels prestained with ethidium bromide. Individual alleles were identified by fragment length and by the corresponding allele-specific primers used.
Ethical aspects
The entire study was approved by the Gabonese Ministry of Health. Febrile children's parents or guardians were informed about the study protocol, either for the comparison of diagnostic tools or for the consecutive molecular analysis. Their oral consent was required prior to enrolment.
Data analysis
Data were entered using Epi info version 2000. The frequency of each allele was estimated over all the alleles detected. Allelic family distribution and the number of genotypes detected in each infected patient were calculated according to the site and the age. The complexity of infection, or the mean number of different parasite genotypes per infected individual, was determined according to the site and the age. Differences between groups were assessed using the chi-square or Fisher’s exact tests for proportions. Continuous variables are presented as medians 25th and 75th percentiles, and were compared using the student’s t-test, Mann Whitney U-test or Kruskal-Wallis test as appropriate. A P-value of less than 0.05 was considered significant.
RESULTS
In total, 168 samples collected from children were analyzed. Children’s median age was 36 (24-60) months. More than 70% (n=121) of the patients were less than 5 years old and 92 (54.8%) were male. The median parasite density was 35,700 (4,501-139,883) parasites/µl; it was 42,700 (4,777-139,755) parasites/µl at Oyem and 29,960 (4,378-139,825) parasites/µl at Owendo (P=0.6).
Allelic family distribution according to site and age
K1, Ro33, and Mad20 alleles were found in 65.5% (n=110), 55.9% (n=94), and 20.8% (n=35) of the isolates, respectively. These proportions did not differ according to the site (Oyem vs Owendo); 65.8% (n=56) vs 65% (n=54) for K1 (P=0.7); 60% (n=51) vs 51.8% (n=43) for Ro33 (P=0.2), and 20% (n=17) vs 21.7% (n=18) for Mad20 (P=0.7).
Depending on the age, the proportions of K1 and Mad20 allelic families tended to be lower in isolates from children less than 5 years old: 62% (n=75) vs 74% (n=35) for K1; 18% (n=22) vs 27% (n=13) for Mad20. Inversely, Ro33 allele frequency tended to be higher in this group (60%; n=72 vs 46.8%; n=22 among the older; P=0.09). The frequency of each allelic family was comparable between both sites (Oyem vs Owendo) in isolates from the youngest children; 65.3% (n=49) vs 56.2% (n=26) for K1, 16% (n=12) vs 22.2% (n=10) for Mad20, and 58.7% (n=44) vs 60.8% (n=28) for Ro33 alleles. Likewise, the proportions of isolates carrying K1 allele among children aged more than 5 years at Oyem (n=7/10) and at Owendo (n=28/37) were not different. In contrast, Ro33 and Mad20 alleles were 1.75 (n=7/10 vs 15/37 at Owendo) to 2.3 (n=5/10 vs 8/37 at Owendo) fold more highly detected at Oyem.
MSP1 allelic diversity and study area
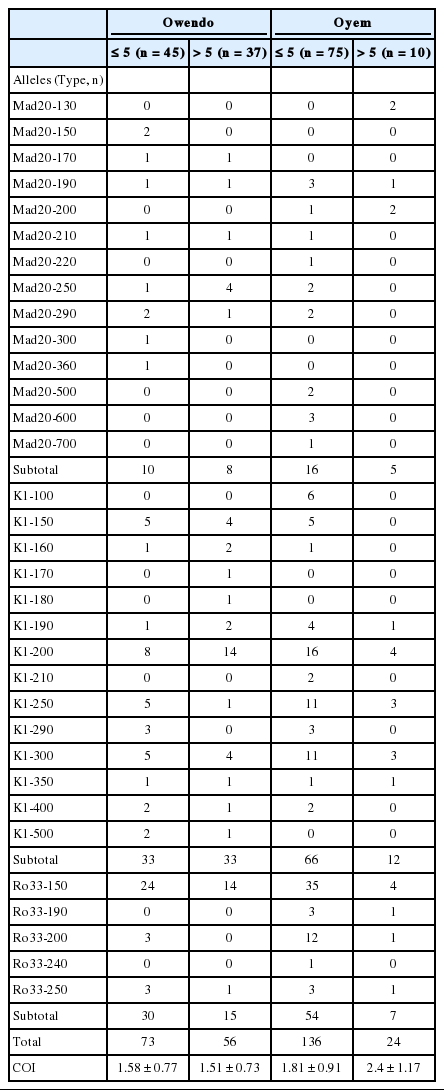
Thirty-three different alleles were detected; 14 K1-type, 14 Mad20-type, and 5 Ro33-type alleles (Fig. 1). A half of these alleles (n=16; 48.5%) was common to the 2 sites; 9 K1-type, 4 Mad20-type, and 3 Ro33-type. Among them, K1-200 and Ro33-150 alleles predominated in isolates from both sites, while among Mad20 alleles the most frequent was Mad-250 at Owendo and Mad-190 at Oyem (Fig. 1).
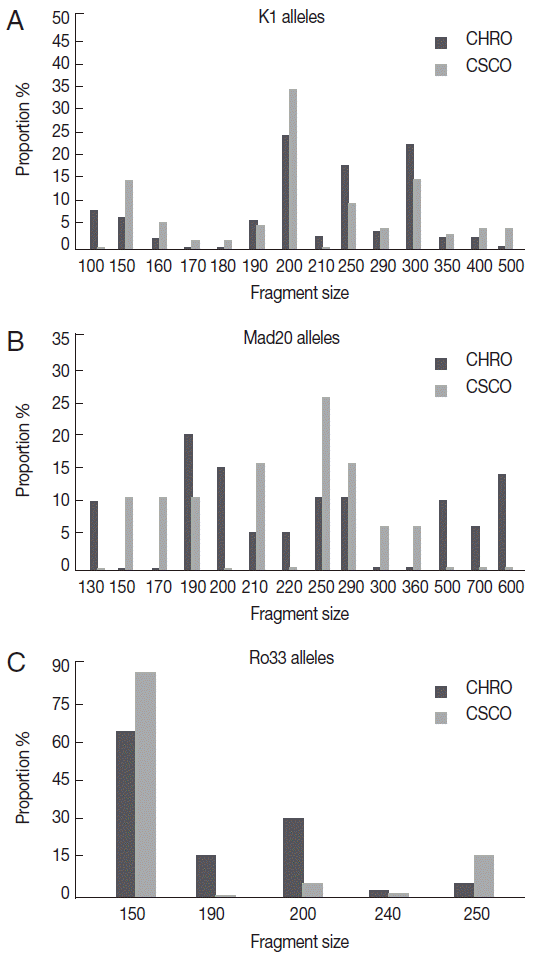
K1, Mad 20, and Ro33 alleles's distribution according to the site. (A) K1 alleles distribution. (B) Mad20 alleles distribution. (C) Ro33 alleles distribution.
The majority of the allele’s site specific has been found at Oyem (n=10/17): 2 Ro33 alleles (Ro33-190 and Ro33-240), 2 K1 alleles (K1-100 and K1-210), and 6 Mad20 (Fig. 1B). No specific Ro33 allele was detected in the urban city of Owendo.
MSP1 allelic diversity and age
In each study site, most of K1, Mad20, and Ro33 alleles were detected in isolates from children aged less than 5 years old; notably Mad20 site specific alleles (Table 1).
Multiples infections
Multiple infections (≥2 genotypes/isolate), with up to 5 different clones per isolate, were found in 50.0% (n=84) of the patients more frequently at Oyem: 57.7% (n=49/85) vs 42.2% (n=35/83) at Owendo (P=0.04). Likewise, isolates with at least 3 different genotypes were predominant at Oyem: 23.5% (n=20/85) vs 10.8% (n=9/83) at Owendo (P=0.03).
In each study site, the frequency of multiple infections was comparable between children; at Oyem 56% (n=42/75) vs 70% (n=7/10) among children aged >5 years old (P=0.6); and at Owendo 43.4% (n=20/46) vs 40.5% (n=15/37) among the youngest and the older, respectively.
The complexity of infection was of 1.72±0.87; higher at Oyem (1.88±0.95 vs 1.55±0.75 at Owendo) (Table 2). Between children ≤5 years old from both sites, the complexity of infection was comparable (P=0.2), while it was the highest among children aged more than 5 years old at Oyem (P=0.03) (Table 2).
DISCUSSION
The present study provides data on P. falciparum genetic diversity from areas in Gabon where this information is lacking, although at least one fifth of the population live there. The 3 msp1 allelic families were found in P. falciparum isolates collected at Oyem and Owendo with a predominance of K1 and Ro33 alleles as previously described in other areas of the country [12,18]. Genetic diversity was the highest within Mad20 and K1 families; 14 different alleles were detected in each family, respectively. A reduced polymorphism was found in Ro33 allelic family as reported elsewhere [19-21]. However, in Mauritania and in Burkina Faso, a highest diversity (>6 alleles) was found within this allelic family [22,23]. A half of the detected alleles, including the most frequent K1-200 and Ro33-150, were common to the parasites from both areas. In contrast, among the Mad20 common alleles, Mad-190 was predominant in the rural area, whereas Mad-250 was prevalent in the urban area. These findings corroborate with those from a study carried out in Papua New Guinea and Tanzania where more than 20% of msp1 alleles were found in the isolates from these 2 countries [19]. Otherwise, the common msp1 alleles were mainly found at Owendo, the urban area, while the specific ones were preponderant at Oyem, in the rural area, underlying a different genetic profile of the parasites between both areas.
Furthermore, these findings suggest a relationship between the highest parasite genetic diversity found at Oyem and the prevalence of confirmed malaria cases in this area. It is accepted that, the majority of malaria episodes are due to a parasite population distinct from the primary infection carried by an individual [24]. The extended allelic diversity found at Oyem would be associated to an increase of the risk of being infected by genetically different parasites and could partly explain the higher frequency of malaria cases among children in this area where malaria prevalence is >40% compared to Owendo. Otherwise, the slow acquisition of immunity to malaria is thought to be due to the need of repeated exposure to a wide diversity of parasite strains. Malaria immunity develops after several malaria clinical episodes, and immunity to common circulating antigens appears more rapidly and should be longlasting. Therefore, the appearance during clinical episodes of parasite genotypes not encountered previously supports a strain-specific component in clinical protective immunity [24].
Site-specific alleles are frequent in the rural area as observed by Soulama et al. [23] in Burkina Faso. Nevertheless, they are detected in lower proportions than common genotypes in both sites, probably due to their “instability” or technical limitations. It has been reported that some parasite genotypes could undergo major fluctuations in density, while others are highly stable [6,25-27].
The rate of polyclonal infections varied according to the site. At the rural area of Oyem the COI and the rate of multiple P. falciparum infections were the highest compared to Owendo and to previous data from other cities [18]. Although, malaria morbidity declined after the deployment of ACTs and bednet in Libreville, Port-Gentil, Franceville and Oyem, malaria prevalence remained the highest in this rural area [28]. The allelic diversity seems to confirm a greater malaria transmission in this city where presumably a significant inoculation of genetically different parasite populations occurs. Indeed, the level of transmission may affect the occurrence of mixed infections; polyclonal infections being more common in intense malaria transmission settings [19,21,23,29]. Moreover, the highest burden of malaria at Oyem could also be due to an absence of the control strategies impact or their insufficient deployment and it can suggest that the genetic profile of circulating parasites cannot be influenced only 3 years after the deployment of new control strategies [30].
The association between age and complexity of infection is not well understood. Some authors did not find any relation between the age and COI as found here. At Oyem, the rural area, the complexity of infection tended to increase with age as reported in a rural area in Burkina Faso [3,21-23,31]. However, an inverse tendency was observed in other endemic areas [32,33]. Moreover, in the present study, the complexity of infection among children aged more than 5 years was lower in the urban city of Owendo compared to that obtained in the rural area of Oyem.
The main limitation of this study is the use of a single marker that has most likely reduced the genetic diversity evaluation of the parasites strains. Nonetheless, previous studies showed an extended genetic diversity of P. falciparum using msp1 gene as well as a correlation between endemicity and the number of msp1 gene distinct alleles [3]. The genetic diversity would probably be more highlighted if the sequence variation had been examined in isolates from both study sites.
A high genetic diversity of P. falciparum isolates from symptomatic children is observed at Oyem, a rural area of Gabon. A half of the detected alleles were common to urban and rural cities. The specific alleles were more frequently found in the rural area although at low frequency. Thus, the proportions of allele’s specific site compared to the common genotypes should be taken into accounts when designing MSP1 or MSP2 malaria vaccine.
Acknowledgements
The authors thank all the outpatients, their parents/guardians, as well as the staff at each of the health facilities, namely, Centre de Santé Communautaire d'Owendo and Centre Hospitalier Régional d'Oyem. They thank all the technicians of the Département de Parasitologie Mycologie et Médecine Tropicale at the Faculty of Medicine, Université des Sciences de la Santé, Gabon.
Notes
We have no conflict of interest related to this work.