DEAD/DExH-Box RNA Helicases in Selected Human Parasites
Article information
Abstract
DEAD/DExH-box RNA helicases catalyze the folding and remodeling of RNA molecules in prokaryotic and eukaryotic cells, as well as in many viruses. They are characterized by the presence of the helicase domain with conserved motifs that are essential for ATP binding and hydrolysis, RNA interaction, and unwinding activities. Large families of DEAD/DExH-box proteins have been described in different organisms, and their role in all molecular processes involving RNA, from transcriptional regulation to mRNA decay, have been described. This review aims to summarize the current knowledge about DEAD/DExH-box proteins in selected protozoan and nematode parasites of medical importance worldwide, such as Plasmodium falciparum, Leishmania spp., Trypanosoma spp., Giardia lamblia, Entamoeba histolytica, and Brugia malayi. We discuss the functional characterization of several proteins in an attempt to understand better the molecular mechanisms involving RNA in these pathogens. The current data also highlight that DEAD/DExH-box RNA helicases might represent feasible drug targets due to their vital role in parasite growth and development.
INTRODUCTION
RNA helicases are remodeling enzymes that catalyze the separation of double-stranded RNA molecules in an energydependent manner in prokaryotic and eukaryotic cells, as well as in many viruses [1]. They belong to the superfamilies 1 and 2 (SF1 and SF2) of helicases. Based on protein sequence, structure, and phylogenetic relationships, SF1 is divided into 3 families (UvrD/Rep, Pif1-like, and Upf1-like), while SF2 includes 9 families (RecQ-like, RecG-like, Rad3/XPD, Ski2-like, T1R, Swi/Snf, RIG-I-like, DEAD-box, and DEAH/RHA) and 1 group (NS3/NPH-II) (Table 1). Of these, Upf1-like, Ski2-like, RIG-I-like, DEAD-box, DEAH/RHA, and NS3/NPH-II families only contain RNA helicases that are frequently referred to as DEAD/DExH or DExD/H proteins [2,3]. DEAD/DExH-box families represent the best and more extensively characterized RNA helicases. These proteins share a -400 amino acid residues catalytic core known as the helicase domain that can be divided into 2 RecA-like domains containing 9 highly conserved motifs (Q, I, Ia, Ib, II, III, IV, V, and VI), as well as GG and QxxR additional motifs. Notably, the 4 amino acids of motif II give their name to DEAD (Asp-Glu-Ala-Asp) and DExH-box (Asp-Glu-x-His) proteins. The helicase domain is flanked by N- and C-terminal regions that are usually not conserved across DEAD/DExH-box proteins but confer specificity for biochemical target and biological function through interaction with other proteins or RNA [3]. Helicase motifs are responsible for the essential biochemical features of DEAD/DExH-box RNA helicases. Q, I, II, V, and VI motifs participate in ATP binding; Ia, GG, Ib, IV, QxxR, and V motifs are involved in RNA recognition, while II, III, V, and VI motifs connect both binding sites to promote RNA duplexes destabilization in an ATPase-dependent reaction (Fig. 1A). These activities likely depend on oligomerization and posttranslational modifications for selected helicases [4,5]. The coordination of these fundamental activities allows DEAD/DExH-box proteins to utilize the free energy change of binding and hydrolyzing a nucleotide triphosphate to dissociate RNA duplexes or displace bound proteins to remodel RNA-protein complexes. Several proteins also can remain bound to RNA (RNA clamping) or promote RNA annealing in an ATP-dependent or independent way. Others function as assembly platforms for larger ribonucleoprotein complexes or can sense bacterial metabolites [6-8].
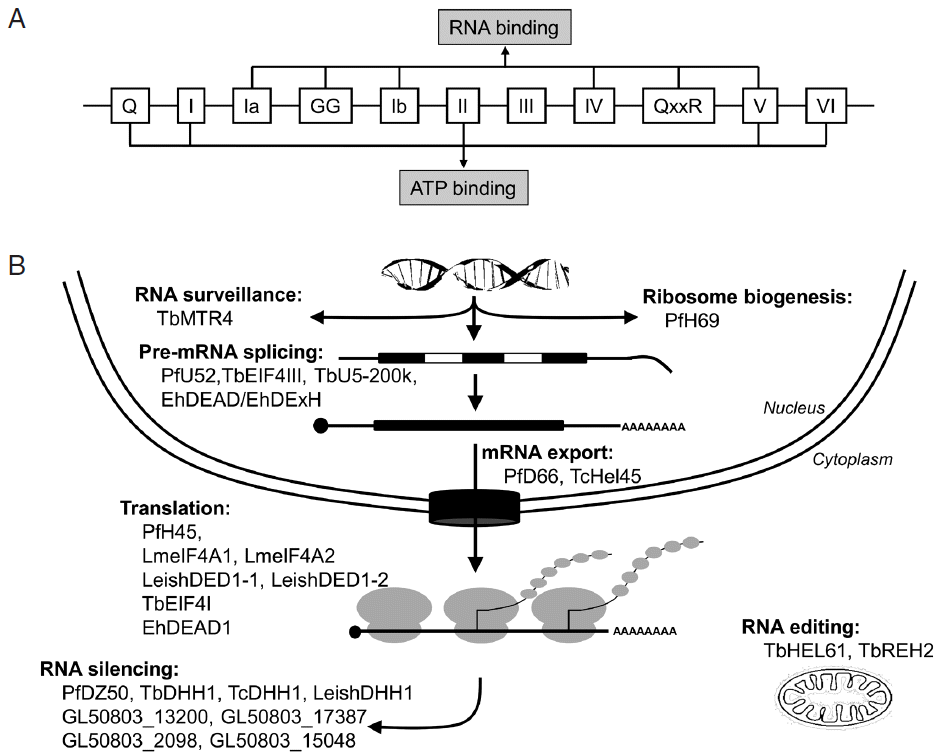
Molecular organization and functions of selected DEAD/DExH-box RNA helicases from human parasites. (A) Schematic representation of the functional helicase domain. (B) Graphical representation of RNA cycle with the main steps involving DEAD/DExH-box RNA helicases from protozoan parasites. In nucleus, these include RNA surveillance, ribosome biogenesis, pre-mRNA splicing, and mRNA export. In cytoplasm, these processes are mRNA translation regulation, RNA silencing, as well as RNA editing in mitochondria.
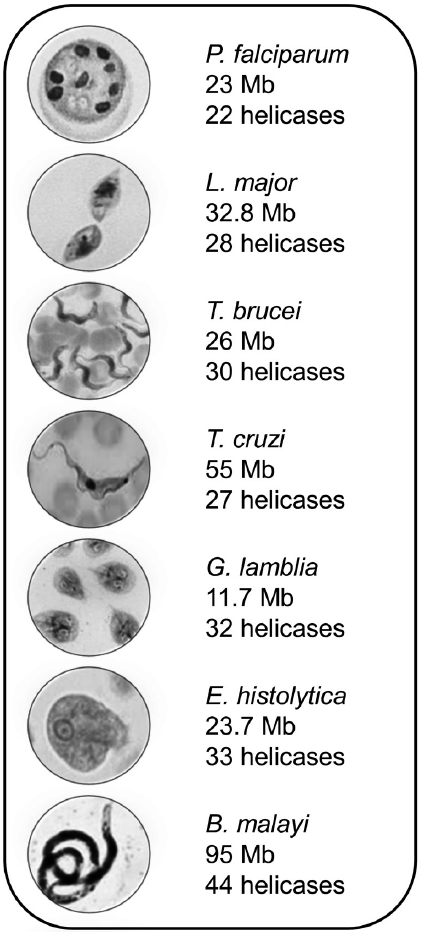
Large RNA helicases families have been described in various organisms, including mammals, plants, yeast, bacteria, and human pathogens, where they participate in many aspects of RNA metabolism, from transcriptional control to mRNA decay, including transcription, pre-mRNA splicing, RNA editing, nucleocytoplasmic transport, translation, ribosome biogenesis, and mRNA degradation, playing an essential role in gene expression regulation [9]. DEAD/DExH-box RNA helicases are therefore biologically relevant molecules required by all living organisms. Indeed, they have been implicated in various human diseases, including viral and other infections, as well as neurological disorders, aging, and cancer [10]. In this review, we summarized the current knowledge about DEAD/DExH-box proteins in selected protozoan and nematode parasites of medical importance worldwide, including Plasmodium falciparum, Leishmania major, Trypanosoma spp., Giardia lamblia, Entamoeba histolytica, and Brugia malayi. Genome-wide studies showed that these organisms possess a large number of DEAD/DExH-box RNA helicases, from 22 in P. falciparum to 44 in B. malayi (Fig. 2). Although the majority of the predicted proteins remain uncharacterized, the functional characterization of several enzymes revealed their relevance in different molecular processes involving RNA, contributing to a better understanding of the biology of these pathogens. Finally, some data also suggested that DEAD/DExH-box proteins could represent potential biochemical targets for anti-parasite treatments.
PROTOZOAN AND NEMATODE PARASITES HAVE LARGE DEAD/DExH-BOX RNA HELICASE FAMILIES
P. falciparum that is transmitted by Anopheles mosquitoes is the protozoan parasite responsible for human malaria. In 2010, more than 2 million cases of malaria and about 660,000 deaths were reported in tropical countries worldwide, mainly in children in Africa, Asia, and South America [11]. P. falciparum genome, 23 megabases (Mb), corresponds to about 5,400 genes distributed on 14 chromosomes, a circular plastid genome, and a mitochondrial genome. Preliminary bioinformatics analyzes indicated that P. falciparum has at least 22 full-length putative DEAD-box helicases genes, some of which being homologs of well-characterized RNA helicases from other organisms [12]. Later, a genome-wide analysis allowed the identification of additional helicase genes. The DEAD-box genes of P. falciparum are unequally distributed on each of the 14 chromosomes and encode proteins whose molecular size varies from 44 to 164 kDa. All P. falciparum proteins contain the conserved helicase domain, but the length and sequence of N- and C-terminal extensions and intervening sequences are variable. Moreover, many Plasmodium proteins present homorepeats of amino acid residues (asparagine, serine, tyrosine, lysine, among others) in long terminal regions, as well as between specific motifs of the helicase domain. The relevance of these sequences for enzyme activity remains unknown [13].
The protozoan parasites Leishmania spp. and Trypanosoma spp. are hemoflagellates of the Trypanosomatidae family. These pathogens are of considerable medical significance and the etiological agents of leishmaniasis, sleeping sickness (African trypanosomiasis), and Chagas disease (American trypanosomiasis). About 12 million individuals are infected with different species of Leishmania, while 0.5 and 10 million people are infected with T. brucei and T. cruzi, respectively [14]. These early divergent eukaryotes have many singular molecular processes, including RNA editing, and trans-splicing, that require specific RNA helicases [15]. The genome of L. major consists of 36 chromosomes that span 32.8 Mb and contain 911 non-coding RNA genes, 39 pseudogenes, and 8,272 protein-coding genes. Remarkably, L. major lacks general transcription factors, and protein-coding genes are organized in polycistronic clusters, indicating that mechanisms regulating RNA polymerase II directed transcription are distinct from those operating in other eukaryotes. L. major has a large number of RNA-binding proteins, which is consistent with the active posttranscriptional regulation of gene expression described in this organism. Notably, initial in silico analyzes revealed the presence of 14 putative RNA helicases [16]. The 26 Mb genome of T. brucei corresponds to 9,068 predicted genes, including about 900 pseudogenes and 1,700 T. brucei-specific genes, distributed over 11 chromosomes. It is characterized by large subtelomeric arrays corresponding to a set of 806 variant surface glycoproteins genes that are used by the parasite to evade the mammalian immune system [17]. The T. cruzi genome contains 55 Mb distributed over 28 chromosomes, with about 12,000 proteincoding genes [18]. A comparative genomic analysis of both Trypanosoma genomes showed that they display high levels of synteny and share a conserved set of approximately 6,200 genes [19]. Recently, Gargantini et al. [20] identified a total of 103, 112, and 113 putative helicases in L. major, T. brucei, and T. cruzi genomes, respectively, from an extensive search in the TriTryp database. Interestingly, most predicted proteins are DEAD/DExH-box helicases with 27-30 members in the 3 species [15].
G. lamblia (also known as G. duodenalis and G. intestinalis) is the protozoan parasite that causes giardiasis in humans and a range of domestic and wild mammals, in developing and developed countries [21]. It has mitochondrial organelles called mitosomes [22] and 2 nuclei that contain 1.2×107 bp of DNA with a GC content of 46% [23]. From BLAST analyzes using the human eIF4A and DHX8 amino acid sequences as DEAD-box and DExH-box helicase prototypes, respectively, Gargantini et al. [20] identified a set of 32 putative RNA helicases, including 22 DEAD-box and 6 DEAH-box proteins. G. lamblia proteins present high sequence similarity with characterized human and yeast homologs, which suggests that they may have a similar function in RNA metabolism. Gene expression assays indicated that 20 DEAD/DExH-box proteins were up-regulated after encystation induction, while 1 DEAH-box gene was down-regulated. On the other hand, 16 RNA helicases were modulated during the antigenic variation process. These data suggested that DEAD/DExH-box RNA helicase may regulate gene expression during adaptive processes in this parasite [23].
E. histolytica is the etiological agent of human amebiasis, which has a worldwide distribution with a higher prevalence in developing countries. This protozoan parasite causes intestinal dysentery and hepatic abscesses that result in 70,000 to 100,000 deaths per year, making it a leading cause of death in humans [24]. E. histolytica genome is 23.7 Mb in size; 9,938 genes with an average size of 1.17 kb have been predicted. Only 25% of genes may be potentially spliced, and 6% contains introns. Genome sequence analysis revealed a variety of metabolic adaptations: reduction or elimination of mitochondrial metabolic pathways, the use of oxidative stress enzymes associated with anaerobic prokaryotes, as well as evidence for lateral gene transfer of bacterial genes [25]. We reported that E. histolytica has an extensive DEAD/DExH-box RNA helicase gene family that is constituted by 20 EhDead and 13 EhDexh-box genes. Phylogenetic analysis and the absence/presence of particular helicase motifs and introns suggest that various clusters of this large family may be the result of gene duplication, mutations, non-coding DNA sequence addition, and gene fusion events [26].
Besides protozoan pathogens, lymph-dwelling nematodes of the superfamily Filarioidea also caused important human diseases designated as lymphatic filariasis. An example of these parasites is the filarial worm B. malayi that is transmitted by mosquitoes. It infects 13 million people in South and Southeast Asia, causing lymphedema or elephantiasis due to obstruction of the lymphatic system by adult worms for 5-15 years [27]. B. malayi genome represents the first nematode genome to be decoded. It corresponds to approximately 95 Mb organized in 5 chromosomes and predicts 11,500 protein encoding genes [28]. Recently, Tuteja et al. [29] carried out a genome-wide computational analysis that allowed the identification of 44 helicases, including 24 DEAD-box helicases and 7 DEAH-box helicases.
PARASITE DEAD/DExH-BOX RNA HELICASE PARTICIPATE IN VARIOUS MOLECULAR EVENTS INVOLVING RNA
The high amino acid sequence homology that predicted parasite RNA helicases share with known homologs in other organisms suggests that they might have the same functions. Indeed, the functional characterization of selected DEAD/DExH-box RNA helicases in protozoan and nematode pathogens revealed that they are essential components of many biochemical events involving RNA, from transcription regulation to mRNA decay, including nuclear RNA surveillance, ribosome biogenesis, pre-mRNA splicing, mRNA export, translation regulation, RNA silencing, and RNA editing (Fig. 1B). Interestingly, the phylogenetic analysis using the maximum-likelihood method [30,31] revealed that several parasite proteins are clustered according to their participation in different steps of RNA metabolism (Fig. 3).
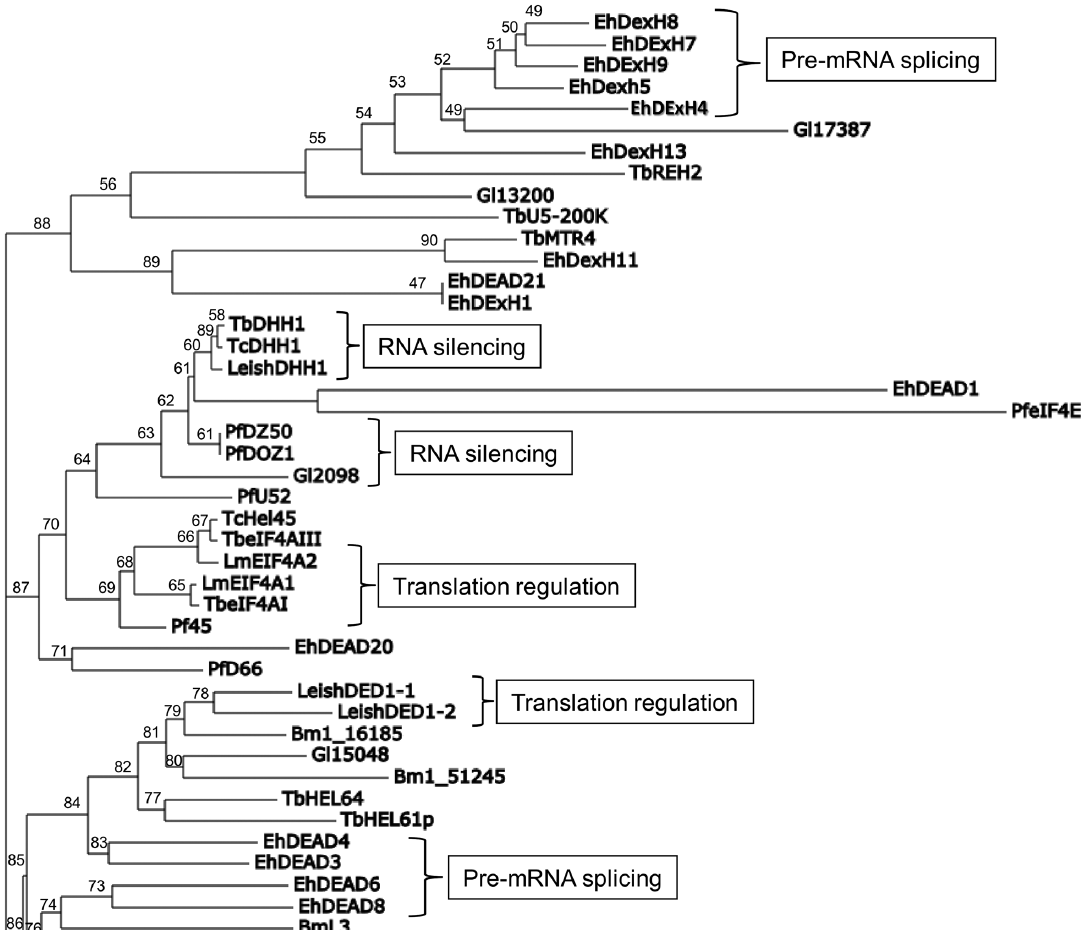
Maximum-likelihood tree of parasite DEAD/DExH-box RNA helicases. This tree was estimated with randomized accelerated maximum-likelihood (RAxML) method based on amino acid sequences of functionally characterized parasites proteins helicases. Support values are given by RAxML bootstrap percentages based on 100 replicates.
Nuclear RNA surveillance
The RNA quality control in the nucleus is an essential mechanism for gene expression. In yeast, it requires coordinated activities of the exosome and the TRAMP complex, which is formed by the interaction of the DExH-box RNA helicase Mtr4p with a non-canonical poly(A) polymerase (either Trf4p or Trf5p), and 1 of 2 RNA-binding proteins (Air1p and Air2p). The TRAMP complex adds a short poly(A) tail to defective RNA molecules, which promotes MTR4-stimulated exosomal degradation [32]. In T. brucei, Cristodero and Clayton [33] identified the nuclear TbMTR4, which presents all the characteristic residues and helicase motifs of homologous DExH-box proteins, and shares 41.5% identity with the yeast RNA helicase Mtr4p. Depletion of TbMTR4 caused parasite growth arrest and rRNA processing alterations; it also produced an increase in RNA polyadenylation. Although the interaction of TbMTR4 with T. brucei homologues of Air1p, Air2p, and Trf4/5p to form a TRAMP complex in trypanosomes remains to be demonstrated, the authors concluded that TbMTR4 is important for deadenylation, which promotes RNA degradation by exosome complex through the polyadenylation-dependent nuclear quality-control pathway [33].
Ribosome biogenesis
Formation of ribosomes involves the synthesis and processing of rRNAs, followed by their assembly with ribosomal proteins, which mainly include AAA-ATPases, GTPases, kinases, and ATP-dependent RNA helicases [34]. In yeast, 1 of these helicases is Has1p that participates in the maturation of 18S rRNA and the formation of the 40S ribosomal subunit [35]. In P. falciparum, Prakash and Tuteja [36] reported the cloning, expression, and characterization of a member of the DEAD-box family designed as PfH69, which is the homolog of yeast Has1p. However, to our knowledge, the role of PfH69 in ribosome biogenesis has not been shown yet. PfH69 lacks the Walker-A-like motif downstream of motif VI, such it has been previously reported for its yeast counterpart. Functional assays revealed that PfH69 is a single-stranded DNA and RNA-dependent ATPase and an RNA binding protein that can unwind DNA and RNA molecules. Surprisingly, besides the conserved helicase domain, the highly variable N-terminal region of the protein also contributes to all activities of PfH69 [36].
Pre-mRNA splicing
In eukaryotic cells, nascent pre-mRNA molecules resulting from transcription are processed in the nucleus before being exported to the cytoplasm and translated. Notably, introns are removed, and exons are joined in a process named splicing that consists of various reactions that are catalyzed by the spliceosome. This complex contains small nuclear ribonucleoproteins (snRNPs) and auxiliary proteins, such as specific DEAD/DExH-box RNA helicases that help to unwind dsRNA fragments [37]. A few years ago, Tuteja [38] reported the detailed computational analysis of 8 predicted Plasmodium RNA helicases that are the homologs of yeast proteins involved in splicing. Among them, only PfU52, which is the homolog of human UAP56 (Sub2p in yeast), has been characterized. ScSub2p/UAP56 is required at multiple steps in splicing and remains associated with the spliced mRNA as part of the exon junction complex (EJC), together with eIF4AIII, another DEAD-box protein. PfU52 has nucleic acid-dependent ATPase, RNA binding, and RNA helicase activities. Mutagenesis assays showed that residues at position 181, 182, and 206 are involved in RNA binding; moreover, ATPase activity depends on nucleic acid binding. Using PfU52 immunodepleted parasite extracts supplemented with recombinant wild-type and mutant PfU52 protein in in vitro splicing reaction, the authors demonstrated that PfU52 RNA binding activity is required for splicing [39].
In T. brucei, 2 DEAD/DExH-box RNA helicases related to splicing events have been described. By genomic searches in T. brucei genome sequence, Dhalia et al. [40] identified a gene that corresponds to a DEAD-box RNA helicase with high homology with eIF4AIII, an EJC component. Congruently, TbEIF4AIII was found in the nucleus. Moreover, the low abundance of TbEIF4AIII, the delayed response to depletion of TbEIF4AIII through RNA interference, and the lack of effects by the dominant negative mutant, indicated that TbEIF4AIII does not play an essential role in protein synthesis. These observations suggest that it may have a more substantial participation in splicing [40]. Recently, Silva et al. [41] reported the T. brucei U5-200K protein, which corresponds to the human U5-200K protein (BRR2 in yeast). This U5 snRNP specific protein is part of the spliceosome, together with the U1, U2, U4, and U6 snRNP particles and non-snRNP proteins [41]. These proteins are DEAH-box helicases with 2 helicase domains, as well as 2 Sec63 domains that seem to unwind the extended U4/U6 duplex during spliceosome catalytic activation [42]. Interestingly, tandem affinity purification assays revealed that T. brucei U5-200K interacts with U5-Cwc21, a novel U5-specific protein that is essential for cis and trans-splicing reactions in trypanosomatids [43]. As expected, T. brucei U5-200K appeared as speckles that concentrate in the nucleus. Notably, U5-200K displays a strong interaction with all U snRNPs, but mainly with U5 snRNP, confirming that it is a U5 snRNP specific protein involved in splicing reactions in T. brucei [41]. In E. histolytica, we also reported 2 proteins with high homology to the U5 snRNP-200 kDa factor, EhDExH1, and EhDExH10. Both predicted proteins present 2 direct repeats of about 750 and 550 amino acids, respectively; each repeat contains 1 helicase domain with only 4 motifs (motifs I, II, III, and VI), as well as the Sec63 motif [26]. Recently, the EhDExH10/U5 snRNP-specific 200 kDa protein, as well as EhDEAD1, EhDEAD3, EhDEAD4, EhDEAD20, EhDEAD6, EhDEAD9, EhDEAD18, EhDExH1, EhDExH4, EhDExH5, EhDExH7, EhDExH8, EhDExH9, EhDExH12, and EhDExH13 helicases, were identified as components of E. histolytica in vivo assembled pre-mRNA splicing complexes, which revealed their participation in the different step of splicing [44].
mRNA export
Nucleocytoplasmic transport of mature mRNA molecules occurs by translocation of mRNA ribonucleoprotein (mRNP) complex through the nuclear pore complexes (NPCs). The mRNP complex formation requires the participation of mRNA and various nuclear proteins, such as RNA export factors, poly(A)-binding protein, nucleoporins, and the DEAD-box protein 5 (Dbp5). Notably, it has been shown that the RNA helicase Dbp5 displaces mRNA bound proteins at the cytoplasmic site of NPCs in both yeast and vertebrates [45]. P. falciparum PfD66 is the homolog of the yeast Dbp5 DEAD-box RNA helicase (DDX19 in human). Sequences similar to PfD66 were also identified in other Plasmodium species and protozoan parasites. The functional characterization of PfD66 revealed that it exhibits MgCl2 and single-stranded DNA dependent ATPase activity. It is a bipolar enzyme that can unwind DNA molecules in both 5´ to 3´ and 3´ to 5´ directions in the presence of the divalent cation Mg2+, Mn2+, or Zn2+. PfD66 is also an ATP-dependent RNA helicase with preference for poly(A) RNA substrates. The relevance of the Q motif and helicase motifs I, Ia, Ib, and II for helicase and ATPase activities was demonstrated using truncated derivatives of PfD66 [46]. The evaluation of a number of DNA intercalating agents on unwinding and ATPase activities of PfD66 evidenced that DAPI, ethidium bromide, netropsin, and nogalamycin are efficient inhibitors, probably because the formation of a compound-DNA complex hampers the movement of the enzyme [47]. To date, the exact role of PfD66 in mRNA export is still unknown. Further experiments are required to understand how it interacts with the other components of mRNA export in P. falciparum.
T. cruzi RNA DEAD-box helicase named Hel45 was identified by comparative genomic analyzes [48] and further characterized by Inoue et al. [49]. The tridimensional modeling of Hel45 indicates that N- and C-terminal domains are linked by a flexible loop and formed a deep cleft, which suggests that it can interact with RNA. Hel45 shares similarity with DBP5/DDX19 and the eukaryotic initiation factor 4AIII (eIF4AIII), both considered as shuttling proteins. Hel45 is a component of mRNP complexes in the cytoplasm but does not associate with polysomes. Moreover, Hel45 was found in the nucleus and clustered around NPCs, which suggests that it acts as a shuttling protein. This hypothesis was confirmed by the expression of a mutant Hel 45 lacking the nuclear export signal motif. Hel45 transport to the cytoplasm is dependent on active transcription and the nuclear mRNA export receptor Mex67, but it is independent of the primary exportin Crm1. Taken all together, these data suggest that Hel45 is related to mRNA export, although its precise function is still unknown.
Translation regulation
After pre-mRNA splicing in the nucleus, several proteins remain bound to mRNA at 20-24 nt upstream of exon-exon junctions to form the EJC. One example is the eukaryotic translation initiation factor 4A (eIF4A). This DEAD-box RNA helicase also known as DDX48 serves as a binding platform for co-ordinating other proteins involved in mRNA metabolism in cytoplasm, including translation and mRNA degradation. Together with eIF4G and eIF4E, eIF4A forms the eIF4F complex that recruits ribosomal subunits to mRNA for subsequent translation [50]. P. falciparum DEAD-box RNA helicase denoted as PfH45 shares high similarity with eIF4A. The recombinant PfH45 protein displays ATP-dependent DNA and RNA helicase activities. It is a bipolar helicase that exhibits both the 3´ to 5´ and 5´ to 3´ directional helicase activities. Remarkably, the inhibition of PfH45 protein was associated with growth inhibition and morphologic deformation of the parasite, demonstrating that PfH45 is essential for P. falciparum survival [51]. The functional characterization of PfH45 showed that its ATPase activity resides in the N-terminus, while the nucleic acid binding activity predominantly resides in the C-terminal of the protein [52]. Searches in L. major databases led to the identification of 2 DEAD-box RNA helicase, LmEIF4A1 (also known as LeIF) and LmEIF4A2, that share 52-59% identity with human eIF4AI [53]. The LmEIF4A1 protein was previously described as an antigen that induces an IL12-mediated Th1 response in peripheral blood mononuclear cells of leishmaniasis patients and acts as a Th1-type natural adjuvant [54]. It is very abundant in L. major promastigotes. Moreover, LmEIF4A1 was found to bind specifically to Leishmania eIF4G homologs, which suggests that it is a component of the eIF4F complex in Leishmania [53]. Additionally, the LeIF DEAD-box RNA helicase of Leishmania infantum is closely related to eIF4A factors. However, complementation assays using an eIF4A-deleted yeast strain (without both essential TIF1 and TIF2 genes encoding eIF4A) revealed that LeIF was not able to substitute for the yeast eIF4A. In contrast, LeIF expression inhibited yeast growth when endogenous eIF4A was expressed from only one of the 2 TIF1 and TIF2 genes. This is probably due to an altered stoichiometry of the translation initiation factors because of the interaction or sequestering of yeast translation factors by LeIF. In accordance with this hypothesis, in vitro binding assays showed that the 25 amino terminal residues of LeIF interact with yeast eIF4G [55]. In T. brucei, the DEAD-box RNA helicase TbEIF4AI shares over 50% identity with the human EIF4A protein. Depletion of TbEIF4AI through RNA interference dramatically reduced protein synthesis and inhibited cell proliferation. Ectopic expression of a dominant negative mutant of TbEIF4AI induced a slow growth phenotype in transfected cells. These data suggested that TbEIF4AI plays a role in translation regulation [40].
The cap binding complex at the 5´ end of mRNA includes the DEAD-box helicase DED1, which also cooperates with the cytoplasmic eIF4F complex that helps the 40S ribosome scan the mRNA from the 5´ end to the AUG start codon [56]. Notably, 2 DEAD-box proteins of Leishmania, denoted as LeishDED1-1 and LeishDED1-2, share high similarity with the yeast DED1 involved in translation initiation. Both proteins seem to emerge from a gene duplication event. Functional characterization showed that both Leishmania DED1 genes were able to complement a mutant yeast strain that fails to express the endogenous DED1. Strikingly, protein synthesis and cell growth arrest were altered only when both LeishDED1-1 and LeishDED1-2 genes were eliminated, indicating that they are functionally redundant, even though they vary in sequence and size. The 2 proteins are expressed in both Leishmania life stages. However, LeishDED1-2 seems to be more abundant in promastigotes of the sandfly midgut, whereas a higher expression of LeishDED1-1 was found in amastigotes inside human host cells. This observation suggests the existence of a partial stage-specific specialization for paralogous proteins. Taken all together, these results indicate that LeishDED1-1 and LeishDED1-2 are prospective regulators of translation initiation in Leishmania [57,58]. The only RNA helicase that has been functionally characterized in E. histolytica is EhDEAD1, which exhibits high homology with the yeast DED1 protein. EhDEAD1 has RNA-dependent ATPase activity and can unwind heteroduplex RNA molecules with 5´ overhangs in an ATPase-dependent manner. It also exhibits a potential single-stranded RNA re-annealing activity, which has been related to the clusters of arginine and glycine residues at the C-terminal in other homologous proteins. RT-PCR assays evidenced that EhDead1 gene is mainly transcribed in cell cycle S phase. Moreover, S to G2/M transition was facilitated when EhDead1 gene expression was inhibited by antisense RNA. These data suggest that EhDEAD1 might participate in S phase and/or posterior cell cycle steps or boundaries in E. histolytica trophozoites. Intriguingly, the ectopic expression of EhDEAD1 was unable to rescue a DED1 defective yeast, which indicates that theses homologous proteins are not functionally conserved between species despite their high sequence homology [59].
RNA silencing
In eukaryotic cells, translationally repressed mRNA are sequestered in 2 kinds of discrete cytoplasmic foci, namely P bodies and stress granules, to be either degraded or stored for later translation. One abundant protein of these cytoplasmic structures is the DEAD-box RNA helicase DDX6, also known as RCK/p54 in mammals and DHH1 in yeast [60]. A few years ago, it was reported that the stabilization and maintenance of translationally dormant mRNAs in the cytoplasm of female gametocytes of P. berghei, a Plasmodium species that infects murine models, depends on the DEAD-box RNA helicase development of zygote inhibited (DOZI), which is the homologue of the human DDX6 [61]. Recently, a DOZI/DDX6 homolog denoted as PfDZ50 was described in P. falciparum, which presents the characteristic DNA and RNA binding, nucleic acid-dependent ATPase and RNA unwinding activities. As expected, PfDZ50 was mainly localized in the cytoplasm of the asexual intraerythrocytic developmental stages of P. falciparum, typically in granular bodies throughout the cytoplasm. Furthermore, PfDZ50 interacts with the recombinant PfeIF4E, the parasite homolog of eIF4E that is part of the eIF4F complex. The full-length PfDZ50 inhibits translation process; notably, translation was restored by about 70% when external PfeIF4E was added. These results suggested that most likely PfDOZI regulates translation by sequestering eIF4E, as it has been described for the p54 RNA helicase of Xenopus, another DDX6 homolog [62].
A DDX6/DHH1 homolog has also been described in T. brucei. The expression of an ATPase-deficient DHH1 mutant protein caused a rapid growth arrest of T. brucei parasites associated with a decrease in polysomes, an increase in P-bodies and a slight decline in average mRNA levels. However, the effect of DHH1 mutant expression on both turnover and translational repression of mRNAs was selective. Notably, the proportion of a specific stabilized mRNA, ISG75, in polysomes was unchanged in the presence of the ATPase-deficient DHH1 mutant, and the ISG75 protein was accumulated, suggesting that DHH1 has a selective role in determining the levels of developmentally regulated mRNAs in insect-stage trypanosomes [63]. In T. cruzi, DHH1 was identified in polysome-independent complexes; it was also located diffusely in the cytoplasm and in cytoplasmic granules whose abundance varies depending on the life cycle status and nutritional conditions. Immunoprecipitation assays revealed that TcDHH1 interacts with proteins of diverse functions, including heat shock proteins, mRNA binding proteins, initiation and elongation translation factors, ribosomal proteins and metabolic proteins. Moreover, TcDHH1-containing complexes of epimastigote also contain mRNA molecules that are mainly expressed in the other forms of the T. cruzi life cycle. These data suggested a role for TcDHH1 in the negative regulation of gene expression in cytoplasmic granules [64]. A homolog of DHH1, the LeishDHH1 protein, has also been identified in Leishmania promastigotes and axenic amastigotes [58].
In Giardia, the antigenic variation process is associated to the up-regulation of the GL50803_13200 and GL50803_17387 proteins, which have high homology with the human DEAH-box helicase RHA (DHX9) involved in remodeling RNA-induced silencing complex to allow dsRNA loading onto this complex, and the GL50803_2098 protein that presents a great homology with the human DDX6 DEAD-box helicase (p54), a general translational repressor that interacts with Ago2 in cytoplasmic P-bodies. In contrast, data revealed the down-regulation of the GL50803_15048 protein, which has a high homology with the Belle (Bel) DEAD-box RNA helicase involved in miRNA and siRNA silencing in Drosophila melanogaster, and 2 related DEAD-box RNA helicases (p68 and p72) that associate with Drosha for miRNA processing in mice [23].
RNA editing
In kinetoplastid protozoa, mitochondrial pre-mRNA molecules can be modified by site-specific insertion and deletion of exclusively uridylate residues to create functional transcripts. This molecular process involves the participation of small RNA molecules known as guide RNAs (gRNAs), a large number of proteins that form the editosome, including RNA helicases [65]. Two mitochondrial DEAD/DExH-box RNA helicases involved in RNA editing have been described in T. brucei, mHEL61p (also named REH1) and REH2. In 1997, Missel et al. [66] reported the identification and characterization of the mitochondrial mHEL61p DEAD-box protein that is similarly expressed in both the insect and bloodstream life cycle stages of the parasite. Importantly, mRNA editing was impaired in the mHEL61 null mutant strain, while ectopic expression of mHEL61 in the knockout cell line was able to recapitulate RNA editing, demonstrating the relevance of mHEL61p for RNA editing [66]. A further functional analysis based on RNAi down-regulation of HEL61p expression experiments revealed that HEL61 is involved in gRNAs displacement either directly by unwinding the gRNA/edited mRNA duplex or indirectly, to allow the 5´ adjacent upstream gRNA to form an anchor duplex with the edited mRNA to initiate another block of editing [67]. On the other hand, the REH2 protein forms unique ribonucleoprotein complexes (RNPs) that have the typical unwinding and gRNA binding activities of helicase enzymes. Moreover, REH2 complexes transiently associate with gRNA and accessory editing factors in mitochondrial RNA-binding complex 1 (MRB1). Based on these results, the authors proposed that REH2-containing structures may be regulating the expression of the mitochondrial genome [68].
DEAD/DExH RNA helicases with unknown functions
HEL64 is a DEAD-box RNA helicase of T. brucei that is essential in insect-stage trypanosomes, since HEL64 double-allele knockout mutants are not viable. The HEL64 of T. brucei has high sequence homology with the nuclear DEAD-box RNA helicase p68 also referred to as DDX5, but its localization in the cytosol of trypanosomes discards the possibility that HEL64 might be a homologue of p68. HEL64 is not the homolog of the conserved translation initiation factor eIF-4A since it is not recognized by an anti-eIF-4A antibody. To date, the HEL64 function remains unknown [69]. Diaz et al. [70] reported the first putative DEAD-box gene RNA helicase gene in T. cruzi by screening in a genomic library. Notably, the predicted HelTc polypeptide contains the conserved motifs characteristic of the DEAD-box protein family. Interestingly, HelTc mRNA is about 8-fold up-regulated in metacyclic trypomastigotes, suggesting that it may function in the differentiation of epimastigotes into infective trypomastigotes. However, the exact role of HEL64 remains unknown [70].
DEAD/DExH-BOX RNA HELICASES AS POTENTIAL THERAPEUTIC TARGETS
Because of their roles in the metabolism of RNA, RNA helicases are essential for parasite biology. Thus, the specific knockdown of BmL3 DEAD-box RNA helicase in B. malayi produced a decrease in motility, viability (97%), and release of microfilariae (81%) from adult females that was related to phenotypic deformities in intrauterine developmental stages. BmL3-helicase inhibition also resulted in death of adult male worms [71,72]. Several RNA helicases have also been shown to be essential for protozoan parasites, such as TbMTR4 [33], TbEIF4A1 [40], TbDHH1 [63], and Hel64 [69] of Trypanosoma spp. Moreover, some predicted parasite RNA helicases present differences in size and domains sequences in comparison with human proteins. For example, Bm1_16185 of B. malayi is slightly larger in size and contains a longer N-terminal region than its human homolog DDX3Y. Bm1_51245 is slightly smaller in size and has a smaller N-terminal region when compared to its human counterpart DDX4; another example is the Bm1_47285 whose helicase domains lacks motif IV [29]. These data suggest that DEAD/DExH-box RNA helicases could be utilized as rational biochemical targets for developing new anti-parasite treatments and overcome the problem of drug resistance. It has been recently published that it is possible to selectively inhibit the activity of the eukaryotic initiation factor (eIF)4A (DDX2), although it has a high similarity with other DExH/D box family members [73]. This knowledge should help to develop new molecules to target parasite DExH/D box family members. Moreover, the detailed characterization of parasite helicases through in silico assays based on molecular dynamics simulation or docking experiments may help to develop inhibitors to specifically block the parasite development.
CONCLUSION
RNA helicases are essential enzymes for many cellular and molecular events that require a transient unwinding of the RNA molecule to provide an appropriate substrate for transcription regulation, nuclear RNA surveillance, ribosome biogenesis, pre-mRNA splicing, mRNA export, translation regulation, RNA silencing, RNA editing, and RNA decay. From the completion of genome sequences for a number of human pathogens, genome wide computational analyses revealed that protozoa and helminthes parasites of medical importance, such as P. falciparum, L. major, Trypanosoma spp., G. lamblia, E. histolytica, and B. malayi, have a large family of DEAD/DExH-box RNA helicases. Even though only a few proteins have been biochemically characterized, their sequence similarity to previously studied helicases was helpful to elucidate the biological role of several parasite enzymes, showing that they are not redundant in function and participate in different regulatory mechanisms in the various life forms of parasites. On the other hand, several RNA helicases might represent feasible drug targets due to their vital role in parasite growth and development. There is no doubt that further detailed studies to characterize function and structural biochemistry of parasite DEAD/DExH-box RNA helicases would extend our knowledge of these proteins and their participation in reactions involving nucleic acids in human parasites. This information would also have an enormous impact for compound design to specifically inhibit parasite enzymes, which could improve the control of these human pathogens.
Acknowledgements
This work was supported by National Polytechnic Institute and Autonomous University of Mexico City, Mexico. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Laurence A. Marchat is supported by COFAA-IPN. Itzel Lopez-Rosas is a postdoc fellowship from CONACyT, Mexico.
Notes
We have no conflict of interest related to this work.