Monitoring of Fasciola Species Contamination in Water Dropwort by cox1 Mitochondrial and ITS-2 rDNA Sequencing Analysis
Article information
Abstract
Fascioliasis, a food-borne trematode zoonosis, is a disease primarily in cattle and sheep and occasionally in humans. Water dropwort (Oenanthe javanica), an aquatic perennial herb, is a common second intermediate host of Fasciola, and the fresh stems and leaves are widely used as a seasoning in the Korean diet. However, no information regarding Fasciola species contamination in water dropwort is available. Here, we collected 500 samples of water dropwort in 3 areas in Korea during February and March 2015, and the water dropwort contamination of Fasciola species was monitored by DNA sequencing analysis of the Fasciola hepatica and Fasciola gigantica specific mitochondrial cytochrome c oxidase subunit 1 (cox1) and nuclear ribosomal internal transcribed spacer 2 (ITS-2). Among the 500 samples assessed, the presence of F. hepatica cox1 and 1TS-2 markers were detected in 2 samples, and F. hepatica contamination was confirmed by sequencing analysis. The nucleotide sequences of cox1 PCR products from the 2 F. hepatica-contaminated samples were 96.5% identical to the F. hepatica cox1 sequences in GenBank, whereas F. gigantica cox1 sequences were 46.8% similar with the sequence detected from the cox1 positive samples. However, F. gigantica cox1 and ITS-2 markers were not detected by PCR in the 500 samples of water dropwort. Collectively, in this survey of the water dropwort contamination with Fasciola species, very low prevalence of F. hepatica contamination was detected in the samples.
Fascioliasis is a zoonosis caused by Fasciola hepatica and Fasciolagigantica, 2 trematode species of the genus Fasciola, prevalent in cattle and emerging as a cause of disease in humans. Humans are infected mainly by ingesting raw water plants that are contaminated with the metacercariae [1]. Several reports have indicated that water plants such as watercress, rice, dandelion, Nasturtium, and Mentha spp. harbor Fasciola metacercariae [2].
Water dropwort (Oenanthe javanica) is a perennial herb with a distinctive aroma and is cultivated in marshy areas of Asia and Australia. The fresh stems and leaves are used as a salad or as a seasoning in soups and stews in Korea [3]. Water dropwort has also been used in Korea as a folk medicine for the treatment of jaundice, hypertension, fever, abdominal pain, leucorrhea, mumps, and urinary difficulty [4]. In a biological hazard analysis of the water dropwort, it was reported that Escherichia coli was detected in samples of the herb collected from water dropwort fields [5]. However, there has been no information on Fasciola species contamination in water dropwort. Here, to obtain basic information regarding Fasciola species contamination in water dropwort in Korea, we collected a total of 500 samples from 3 areas, and evaluated Fasciola species contamination by mitochondrial cytochrome c oxidase subunit 1 (cox1) and nuclear ribosomal internal transcribed spacer 2 (ITS-2) DNA sequencing analysis.
Water dropwort samples were obtained between February and March 2015. A total of 500 samples were collected, and the lower parts of water dropwort was initially examined using a stereomicroscope (×10 magnification, Zeiss, Oberkochen, Germany). Next, we determined the presence of cox1 and ITS-2 genes of Fasciola species in each sample using PCR amplification. Briefly, the surface of the lower 20 cm of the water dropwort stem was peeled using a sterile scalpel, and genomic DNA was isolated using a G-DEX™ genomic DNA extraction kit (iNtRON Biotechnology, Seoul, Korea) according to the manufacturer’s instructions. Genomic DNA isolated from an adult F. hepatica worm (Prof. Sung-Jong Hong, Chung-Ang University, kindly provided) and adult F. gigantica worm (Prof. Keeseon S. Eom and Hyeong-Kyu Jeon, Chungbuk National University, kindly provided) were used as a positive control. The primers used for PCR amplification are listed in Table 1. The PCR mixture for the PCR amplification contained 5 μl genomic DNA, 3 μl each of forward and reverse primers, 4 μl dNTP, 5 μl 10× Ex Taq buffer, 0.25 μl Ex Taq polymerase, and 29.75 μl DDW. PCR assays were performed with an initial denaturation step of 94˚C for 30 sec, followed by 30 cycles of denaturation at 98˚C for 10 sec, annealing at 60˚C for 30 sec, and extension at 72˚C for 30 sec, followed by 1 cycle at 72˚C for 10 min and a final hold at 4˚C. Amplifications were generated using a TaKaRa PCR Thermal Cycler (Takara Bio Inc., Otus, Japan). Agarose gel electrophoresis (1.5%) with ethidium bromide staining was used to visualize the ITS-2 PCR products.
Furthermore, to identify the sequence of the PCR products from Fasciola-contaminated water dropwort, we purified the PCR products. Briefly, after electrophoretic separation, the cox1 and ITS-2 PCR products were clearly delineated and sequenced directly by SolGent (Daejeon, Korea). The sequence of PCR products were compared with the complete cox1 and ITS-2 sequences of F. hepatica obtained from GenBank (accession no. GU112476.1 and AJ272053.1, respectively) using Clone Manager software (Sci-Ed Software, Cary, North Carolina, USA). Also, the sequence of PCR products were compared with the complete cox1 and ITS-2 sequences of F. gigantica obtained from GenBank (accession no. ab983838.1 and EU260059.1, respectively).
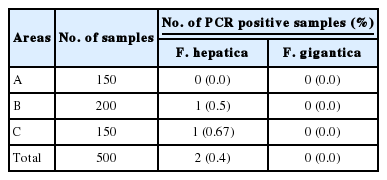
We tried to detect the metacercariae of Fasciola species from the surface of water dropwort using a stereomicroscope. However, metacercariae were not observed at the stack of water dropwort. Next, we performed PCR analysis on the 500 specimens of water dropwort to monitor Fasciola species contamination. Among the 500 specimens collected from 3 areas, cox1 and ITS-2 bands of F. hepatica were detected in 2 specimens (Table 2; Figs. 1, 2), which were exactly consistent with the PCR amplification of the positive control, adult F. hepatica DNA (283 bp for cox1 and 346 bp for ITS-2). We also performed PCR analysis to evaluate F. gigantica contamination of water dropwort using F. gignatica cox1 and ITS-2 gene primers. However, the F. gigantica cox1 and ITS-2 markers were not detected by PCR using 500 water dropwort (data not shown). To confirm whether the positive PCR products were real F. hepatica, the complete DNA sequence of cox1 and ITS-2 PCR products were compared with those of GenBank. The nucleotide sequences of the cox1 PCR products from the 2 F. hepatica-contaminated samples were 96.5% identical to the F. hepatica cox1 sequences of GenBank (accession no. GU112476.1; Fig. 3), whereas F. gigantica cox1 sequences were 46.8% similar with cox1 positive samples. The ITS-2 sequences of 2 PCR positive samples were 100% identical to those of GenBank (accession no. aj272053.1) and positive control sample (adult F. hepatica worm); however, F. giganica ITS-2 sequences were 97.5% identical to those of ITS-2 positive PCR samples (Fig. 4). Thus, Fasciola species PCR positive samples were confirmed to be F. hepatica, and the overall prevalence of F. hepatica infection in water dropwort was 0.4%, ranging from 0.0% to 0.67% depending on the collection area.
Results for the detection of the cox1 and ITS-2 genes of F. hepatica or F. gigantica from water dropwort by PCR

Agarose gel electrophoresis of PCR products containing the mitochondrial cytochrome c oxidase subunit 1 (cox1) marker of Fasciola hepatica. M, 100 bp marker; lane 1, positive control (adult F. hepatica worm); lanes 2-7, F. hepatica negative samples of water dropwort; lane 8; No. 11 F. hepatica positive sample of water dropwort, lane 9; No. 18 F. hepatica positive sample of water dropwort.

Agarose gel electrophoresis of PCR products containing the nuclear ribosomal internal transcribed spacer 2 (ITS-2) marker of F. hepatica. M, 100 bp marker; lane 1, positive control (adult F. hepatica worm); lanes 2-7, F. hepatica negative samples of water dropwort; lane 8; No. 11 F. hepatica positive sample of water dropwort, lane 9; No. 18 F. hepatica positive sample of water dropwort.

F. hepatica cox1 nucleotide sequences of 2 positive samples obtained from PCR products compared with a GenBank sequence (accession no. GU112476.1). Base homologies are indicated by a dot (·); base changes are shown in orange. Fh COX1 GU11, F. hepatica cox1 GenBank sequence (accession no. GU112476.1); No. 1, positive control (adult F. hepatica worm); No. 11, No. 11 F. hepatica positive sample of water dropwort; No. 18, No. 18 F. hepatica positive sample of water dropwort; Fg COX1 AB98, F. gigantica cox1 GenBank sequence (accession no. AB983838.1).

F. hepatica ITS-2 nucleotide sequences of 2 positive samples obtained from PCR products compared with a GenBank sequence (accession no. AJ272053.1). Base homologies are indicated by a dot (·); base changes are shown in orange. Fh ITS-2 AJ27, F. hepatica ITS-2 GenBank sequence (accession no. AJ272053.1); No. 1, positive control (adult F. hepatica worm); No. 11, No. 11 F. hepatica positive sample of water dropwort; No. 18, No. 18 F. hepatica positive sample of water dropwort; Fg ITS-2 EU26, F. gigantica ITS-2 GenBank sequence (accession no. EU260059.1).
Fascioliasis in animals and humans is caused by F. hepatica and F. gigantica. It is difficult to accurately discriminate between 2 species because their size varies depending on the age of the fluke and species of the host [6-8]. PCR technology and DNA sequencing techniques facilitate species identification, clarification of strains, and genetic populations. Genes in the mitochondrial and nuclear DNA (the genes encoding ribosomal RNAs) have been used as marker(s) in population genetics and phylogeny for fasciolid classification [6-8]. The prevalence of fascioliasis was greatly reduced in the 2000s in Korea. However, human cases of F. hepatica infection have been continuously reported [9,10]. Humans and cattle are most commonly infected by ingestion of water plants contaminated with encysted metacercariae. Water dropwort is one of the major sources of F. hepatica infection in Korea [2,9]. In this study, the overall prevalence of F. hepatica infection in water dropwort was 0.4%, which was much lower than that of snails in water dropwort fields in Korea [11]. Moreover, the prevalence in this study was lower than that in watercress in France (1.2-2.4% annually) [12]. Sources of F. hepatica contamination in agricultural products include soil, feces, irrigation water, inadequately composted manure, wild and domestic animals, dirty equipment, and human handling [13]. Differences in prevalence may be induced by various factors such as host distribution, locality, and environmental conditions. In this study, we used the repetitive DNA sequences of cox1 and ITS-2 regions specific for F. hepatica or F. gigantica to identify the species of genus Fasciola because these genes were used efficiently to identify liver fluke species collected from various hosts and geographic regions [6-8]. From this study, F. hepatica cox1 and ITS-2 DNA were detected at 2 samples among 500 samples, but not F. gigantica contamination. These results were further confirmed by sequence analysis of positive PCR products in comparison to cox1 and ITS-2 gene sequences of F. hepatica and F. gigantica.
Taken together, of 500 water dropwort samples, 2 water dropwort samples displayed the DNA bands of F. hepatica via PCR, and these findings were confirmed by sequencing analysis. This is the first study regarding parasitological examination of Fasciola species in water plants in Korea, suggesting that we need to improve the biosafety of aquatic plants during the preand postharvest periods.
Acknowledgements
The present research has been supported by Rural Development Administration, the Republic of Korea (project no. PJ01085903).
Notes
We have no conflict of interest related to this work.
