Prevalence of Anaplasma and Bartonella spp. in Ticks Collected from Korean Water Deer (Hydropotes inermis argyropus)
Article information
Abstract
Deer serve as reservoirs of tick-borne pathogens that impact on medical and veterinary health worldwide. In the Republic of Korea, the population of Korean water deer (KWD, Hydropotes inermis argyropus) has greatly increased from 1982 to 2011, in part, as a result of reforestation programs established following the Korean War when much of the land was barren of trees. Eighty seven Haemaphysalis flava, 228 Haemaphysalis longicornis, 8 Ixodes nipponensis, and 40 Ixodes persulcatus (21 larvae, 114 nymphs, and 228 adults) were collected from 27 out of 70 KWD. A total of 89/363 ticks (266 pools, 24.5% minimum infection rate) and 5 (1.4%) fed ticks were positive for Anaplasma phagocytophilum using nested PCR targeting the 16S rRNA and groEL genes, respectively. The 16S rRNA gene fragment sequences of 88/89 (98.9%) of positive samples for A. phagocytophilum corresponded to previously described gene sequences from KWD spleen tissues. The 16S rRNA gene fragment sequences of 20/363 (5.5%) of the ticks were positive for A. bovis and were identical to previously reported sequences. Using the ITS specific nested PCR, 11/363 (3.0%) of the ticks were positive for Bartonella spp. This is the first report of Anaplasma and Bartonella spp. detected in ticks collected from KWD, suggesting that ticks are vectors of Anaplasma and Bartonella spp. between reservoir hosts in natural surroundings.
The close association of wild animals and birds and their associated arthropod vectors with human populations and domestic livestock, travel, international trading of livestock, and illegal wildlife trading assist in the spread of infectious diseases worldwide [1]. Deer serve as reservoirs of several vector-borne pathogens that impact on human and veterinary health throughout much of the world, including the Republic of Korea (ROK) [2-6]. In the ROK, the Korean water deer (KWD, Hydropotes inermis argyropus) populations have rapidly increased from 1982 to 2011, in part, as a result of reforestation programs and the absence of predators and prohibition of hunting [7]. Consequently, KWD are often observed grazing on vegetation near/in rural villages, agricultural areas (e.g., rice paddies), and even in metropolitan cities in close association with human populations, livestock, and poultry farms. A previous survey identified a high prevalence of Anaplasma phagocytophilum, Anaplasma bovis, and Bartonella grahamii in KWD spleen tissues, suggesting that KWD are reservoirs of these pathogens [8-9].
A. phagocytophilum is the causative agent of human granulocytic anaplasmosis and transmitted mainly by Ixodes ticks in many parts of the world [10]. In the ROK, A. phagocytophilum was identified by PCR in deer, rodents, and ticks belonging to the genera Haemaphysalis and Ixodes that were collected by tick drag and from migratory birds [8,11,12]. B. grahamii was first isolated from rodents in the United Kingdom and is transmitted mainly by Ctenocephalides nobilis nobilis, a flea often associated with rodents [14,15]. However, recently Bartonella spp. have been detected in several tick species by PCR in other parts of the world [16]. In the ROK, Bartonella spp. have been detected in ticks, e.g., Haemaphysalis flava, Haemaphysalis longicornis, Ixodes nipponensis, and Ixodes turdus, collected by tick drag and from migratory birds [11,13].
The purpose of this study was to identify tick-borne pathogens, e.g., A. phagocytophilum and Bartonella spp., in ticks that were collected from KWD that serve as suspected natural reservoirs of tickborne pathogens and pose serious vector-borne disease threats to wildlife, domestic animals, and human populations.
The Conservation Genome Resource Bank for Korean Wildlife examined KWD carcasses in the ROK from March 2008-May 2009. A total of 27/70 (38.6%) of the KWD carcasses were positive for ticks. Ticks were removed and placed individually in cryovials containing 70% ethyl alcohol and labeled with a unique identification number that corresponded to the KWD collection data. Ticks were provided to the 5th Medical Detachment, 168th Multifunctional Medical Battalion, 65th Medical Brigade, Yongsan US Army Garrison, Seoul, where they were identified [17] and pooled according to tick species, life stages, sex (adults), and KWD collection numbers and then provided to the Laboratory of Veterinary Internal Medicine, Seoul National University where they were tested for selected pathogens.
Individuals and pools of ticks were homogenized mechanically using a Beadbeater TissueLyser II (QIAGEN, Hilden, Germany) with lysis buffer, proteinase K, and 5 mm stainless steel beads (30 frequencies/s for 5 min), followed by incubation at 56˚C overnight and then centrifugation at 12,000 g for 10 min at 4˚C. The genomic DNA extraction was performed using DNeasy® Tissue Kits (QIAGEN) according to the manufacturer’s instructions. PCR and nested-PCR were performed using generic-specific primer sets for Bartonella and Ehrlichia/Anaplasma, and species-specific primers for A. phagocytophilum, A. bovis, and B. grahamii as described by Kang et al. [13].
PCR products were purified using QIAquick Gel Extraction kits (QIAGEN) and were cloned with pGEM®-T Easy Vectors (Promega Corp.). Plasmid DNA for sequencing was purified using the Wizard® Plus SV Minipreps DNA Purification System (Promega Corp.) according to the manufacturer’s instructions. Purified recombinant plasmid DNA was sequenced by dideoxy termination with an automatic sequencer (ABI 3730xl capillary DNA sequencer, Foster City, California, USA). The DNA sequences were evaluated with Chromas software (Ver 2.33), aligned using Clustal X (Ver 2.1), and then examined with a similarity matrix. The phylogenetic trees of nucleotide sequences were developed using the MEGA6 program. GenBank accession numbers for the 16S ribosomal (r) RNA and ITS region sequences and specific genospecies sequences related to bacterial pathogens for sequence comparisons are shown in Figs. 1, 2.
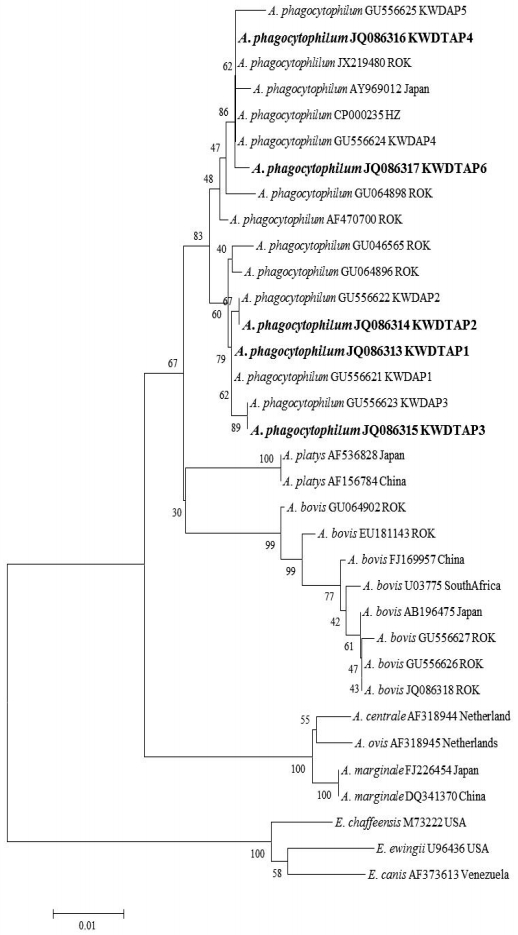
Phylogenetic relationships for Anaplasma phagocytophilum (bold letters) detected from ticks collected from Korean water deer and Anaplasma and Ehrlichia species based on partial nucleotide sequences of 16S rRNA gene fragments (926 bp). The neighbor-joining method was used for constructing the phylogenetic tree. The numbers at the nodes are the proportions of 1,000 bootstrap iterations that support the topology shown.
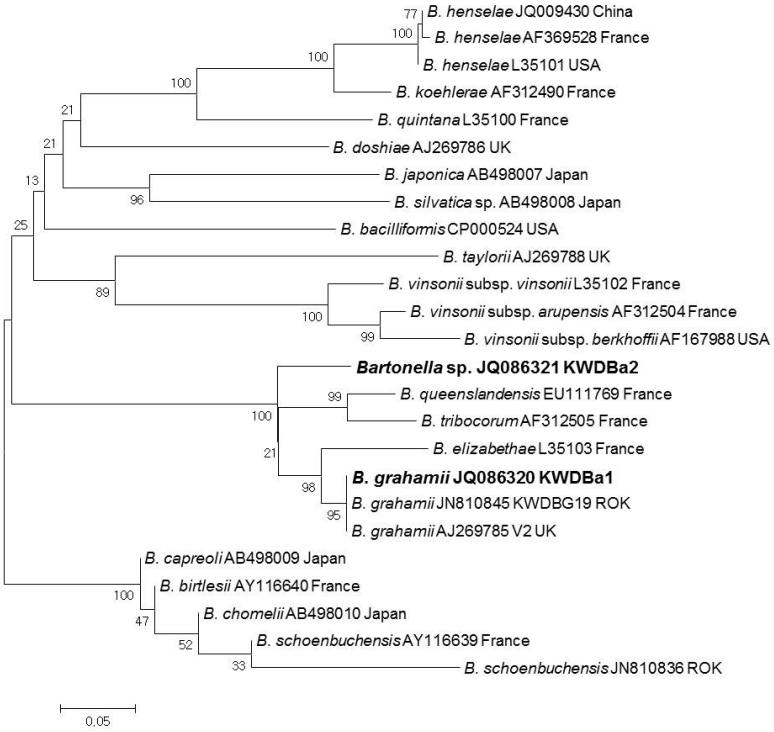
Phylogenetic relationships for Bartonella grahamii and Bartonella spp. (bold letters) detected from ticks collected from Korean water deer and Bartonella spp. based on partial nucleotide sequences of ITS gene fragments (484 bp). The neighbor-joining method was used for constructing the phylogenetic tree. The numbers at the nodes are the proportions of 1,000 bootstrap iterations that support the topology shown.
A total of 363 (21 larvae, 114 nymphs, and 228 adults) fed ticks belonging to 2 genera and 4 species, H. flava (n=87), H. longicornis (n=228), I. nipponensis (n=8), and I. persulcatus (n=40), were collected from 27/70 (38.6%) KWD carcasses (Table 1). The minimum infection rate of A. phagocytophilum was 24.5% (89/363 ticks, 266 pools) for ticks using nested PCR targeting the 16S rRNA gene (Table 1). The 16S rRNA sequences (n=89) of A. phagocytophilum were classified into 5 genotypes that showed 98.7-100% similarity to the A. phagocytophilum HZ strain isolated from a human (CP000235) (Fig. 1). Four of 5 genotypes (KWDTAP1, 2, 3, and 4) corresponded to previously detected 16S rRNA sequences of A. phagocytophilum (KWDAP1, 2, 3, and 4) from KWD spleen tissues [8]. In addition, the 5/363 (1.4%) amino acid sequences of A. phagocytophilum groEL (KWDTAPg, JQ086319) obtained from 2 H. flava, 2 I. nipponensis, and 1 I. persulcatus adult ticks were identical to each other and corresponded to the amino acid sequence of A. phagocytophilum groEL (ADO34908) that was previously reported from KWD spleen tissue. All A. bovis 16S rRNA gene sequences (JQ086318) from ticks were identical and corresponded to the A. bovis sequence (GU556626) previously identified from KWD spleen tissues. A total of 13 (3.6%) of the ticks demonstrated dual infections with A. phagocytophilum and A. bovis.
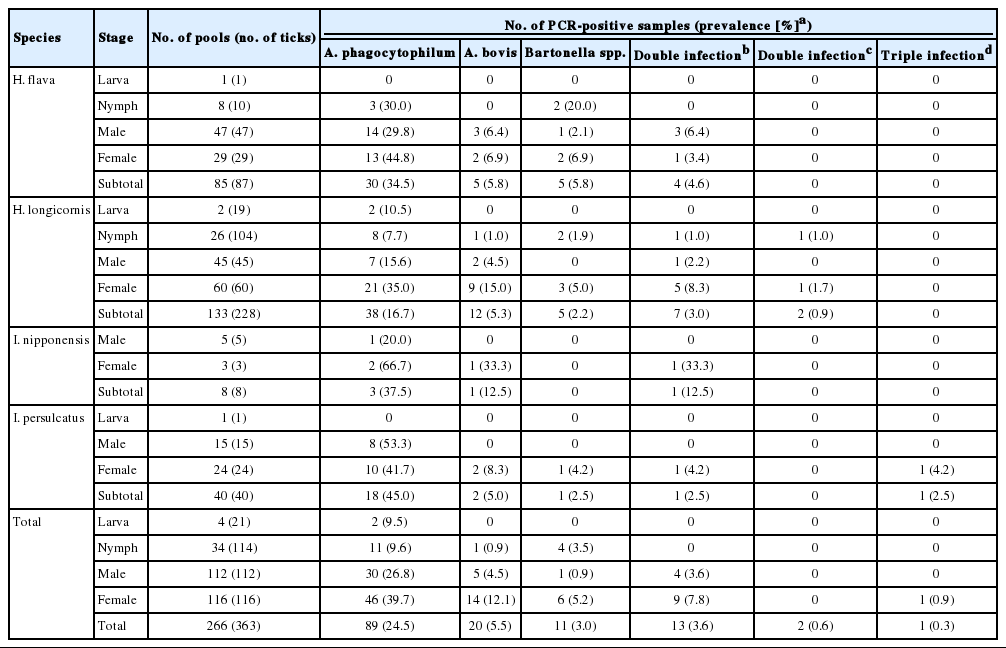
Prevalence of tick-borne pathogens in fed ticks collected from Korean water deer (Hydropotes inermis argyropus) in the ROK using 16S rRNA primer sets
Using the ITS specific nested PCR, a total of 11 Bartonella sequences were classified into 2 genotypes (KWD-Ba1 and KWDBa2) (Fig. 2). KWDBa1 (JQ086210, n=4) was identified as B. grahamii V2 strain (AJ269785) and corresponded to the previous ITS sequence (JN810845) identified from KWD spleen tissues. KWDBa2 (JQ086321, n=7), which showed 92.7% similarity to B. grahamii V2 strain (AJ269785), was identified as Bartonella spp. (Fig. 2). In addition, 2 (0.6%) adult ticks each demonstrated double infections with A. phagocytophilum and Bartonella spp., while 1 (0.3%) individual adult tick demonstrated triple infections with A. phagocytophilum, A. bovis, and Bartonella spp.
KWD and the Korean spotted deer (Cervus nippon mantchuricus) are strongly suspected as reservoirs of Anaplasma and Bartonella species in the ROK [6,11,13]. This study supports evidence that ticks become infected while blood feeding on deer and are vectors of Anaplasma and Bartonella species that are present in deer populations in the ROK. However, there is insufficient correlation of Anaplasma and Bartonella spp. transmission between KWD and blood feeding ticks since some attached ticks collected from A. phagocytophilum positive KWD were negative for A. phagocytophilum, while attached ticks collected from negative KWD were positive for A. phagocytophilum. These results indicate that blood feeding does not always accompany ingestion of these vector-borne pathogens.
Bartonella spp. are mainly transmitted by fleas, but recently Bartonella DNA has been detected in unfed Haemaphysalis and Ixodes spp. collected by tick drag and from partially fed and recently attached ticks collected from migratory birds in the ROK [11,13]. Molecular evidence supports the potential transmission of Bartonella spp. since it is transmitted by transstadial transmission as indicated by positive unfed ticks that were collected by tick drag. Tick bites among ROK populations are largely unreported with an average of only 40 tick bites annually, while there were 55 cases of severe fever with thrombocytopenia syndrome reported for 2014 [18]. Therefore, the potential risk associated with the transmission of A. phagocytophilum and Bartonella spp. in ticks that fed on infected KWD to ROK civilian and military populations and livestock is unknown.
In summary, evidence supports the maintenance of A. phagocytophilum, Bartonella spp. and other tick-borne pathogens in KWD and associated ticks that pose potential serious threats to medical and veterinary health in the ROK.
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (project no. PJ010092)”, Rural Development Administration, the Republic of Korea. This research was also supported by a National Research Foundation of Korea Grant funded by the Korean Government (2011-0015349), the Public Health Command District-Korea, and the Armed Forces Health Surveillance Branch, Global Emerging Infections Surveillance and Response System (AFHSC-GEIS), Silver Spring, Maryland, USA.
Disclosure: The opinions expressed in this article are those of the authors and do not reflect official policy or positions of the US Department of the Army, the US Department of Defense, or the US Government.
Notes
We have no conflict of interest related to this work.