Identification of Protein Arginine Methyltransferase 5 as a Regulator for Encystation of Acanthamoeba
Article information
Abstract
Encystation is an essential process for Acanthamoeba survival under nutrient-limiting conditions and exposure to drugs. The expression of several genes has been observed to increase or decrease during encystation. Epigenetic processes involved in regulation of gene expression have been shown to play a role in several pathogenic parasites. In the present study, we identified the protein arginine methyltransferase 5 (PRMT5), a known epigenetic regulator, in Acanthamoeba castellanii. PRMT5 of A. castellanii (AcPRMT5) contained domains found in S-adenosylmethionine-dependent methyltransferases and in PRMT5 arginine-N-methyltransferase. Expression levels of AcPRMT5 were increased during encystation of A. castellanii. The EGFP-PRMT5 fusion protein was mainly localized in the nucleus of trophozoites. A. castellanii transfected with siRNA designed against AcPRMT5 failed to form mature cysts. The findings of this study lead to a better understanding of epigenetic mechanisms behind the regulation of encystation in cyst-forming pathogenic protozoa.
INTRODUCTION
Acanthamoeba is an opportunistic protozoan pathogen that can cause granulomatous encephalitis and keratitis [1]. Under harsh conditions or chemotherapeutic drugs, Acanthamoeba transforms into a resistant cyst form, causing significant problems in the treatment of Acanthamoeba infections [2]. Recently, several encystation-mediating factors have been found to inhibit the encystation of Acanthamoeba [3-9]. However, the regulation mechanisms of encystation mediating factors are, to our knowledge, still unknown.
Epigenetics studies the molecular mechanisms that control gene expression and is currently one of the most important fields in science. Epigenetics refers to heritable changes in gene expression that do not involve a change to the DNA sequence [10]. In higher eukaryotes, the study of epigenetic mechanisms responsible for various gene expression patterns has been widely used. Epigenetics is associated to processes such as gene and microRNA expression, DNA-protein interactions, suppression of transposable element mobility, cellular differentiation, embryogenesis, X-chromosome inactivation, and genomic imprinting [11]. Epigenetic modifications can be grouped into 3 main categories: DNA methylation, histone modifications, and nucleosome positioning [11].
In yeast and humans, the posttranslational methylation of arginine residues in proteins affects myriad cellular processes, including transcription, RNA processing, DNA replication and repair, and signal transduction [12]. The protein arginine methyltransferase (PRMT) has been reported to catalyze these reactions. PRMTs have been identified in animals, fungi, plants, and protozoa. In parasitic protozoa, PRMTs have been identified in Trichomonas vaginalis, Entamoeba histolytica, Trypanosoma brucei, Plasmodium falciparum, and Toxoplasma gondii [12]. In addition to PRMT1 and PRMT5, cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) have been characterized in E. histolytica [13]. However, to our knowledge, the presence of epigenetic regulation in Acanthamoeba has not been reported yet.
In order to understand epigenetic regulation in Acanthamoeba, we analyzed the cDNA library of Acanthamoeba castellanii, which contains several genes with a methyltransferase domain. Among these sets, the homologue gene of PRMT5 (GenBank accession no. KT345169) was found. In this study, we demonstrate the regulatory role of protein arginine methyltransferase 5 (PRMT5) during encystation in A. castellanii.
MATERIALS AND METHODS
Amoeba cultivation and encystation
A. castellanii Castellani was obtained from the American Type Culture Collection (ATCC 30011). A. castellanii trophozoites were axenically cultured in PYG medium that contains proteose peptone 0.75% (w/v), yeast extract 0.75% (w/v), and glucose 1.5% (w/v) at 25˚C in a Sanyo incubator (San Diego, California, USA). A. castellanii encystation was induced in an encystation media (0.1 M KCl, 0.008 M MgSO4, 0.0004 M CaCl2, and 0.02 M 2-amino-2-methyl-1,3-propanediol, pH 9.0) for 3 days [14]. Mature cysts were counted under a light microscope after treating them with 0.5% SDS, and encystation ratios were calculated [15].
Gene expression analysis based on real-time PCR
Total RNA was purified using TRIzol reagent (Gibco BRL, Rockville, Maryland, USA), and cDNA synthesis was conducted using a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Hanover, Indiana, USA). Real-time PCR was performed using the ABI PRISM® 7000 sequence detection system (Applied Biosystems, Foster City, California, USA), using the default thermocycler program for sense and antisense primers (sense 5´-TTGACTACTCGGCTCTTCTGCC and antisense 5´-TCGATGTCTTTCACCAGCAGG for protein arginine methyltransferase 5). All reaction mixtures used Sybr Premix Ex Taq (Takara, Otsu, Shiga, Japan). The 18S ribosomal DNA was used as the reference gene [7]. Real-time PCR was performed to determine relative gene expression using the 2ΔΔCT method [16], and experiments were performed in triplicate.
Stable transfection
To investigate the intracellular localization of protein arginine methyltransferase 5, the gene was cloned into the pGAPDH vector using enhanced green fluorescent protein (EGFP) as a marker. This plasmid was transfected into live cells of A. castellanii. Following a previously described method, transient transfection was performed using the SuperFect transfection reagent (Qiagen, Hilden, Germany) [17]. Transfected cells were transferred to 50 μg neomycin-containing G418 media and grown over several passages.
Confocal microscopy
The LSM 5 EXCITER Scalable confocal system (ZEISS, Hamburg, Germany) was used to identify and observe amoebae expressing EGFP. EGFP- or DAPI (4´,6-diamidino-2-phenylindole)-mediated fluorescence was performed using bandpass filters covering excitation and emission wavelengths of 500 to 530 nm and 360 to 460 nm, respectively.
Gene silencing
Small interfering RNA (siRNA) targeting protein arginine methyltransferase 5 of A. castellanii (AcPRMT5) was synthesized by Sigma-Proligo (Boulder, Colorado, USA), based on its cDNA sequence. The siRNA duplex with sense (5´-CAGCAAUUGCGCAGUAUCAdTdT) and anti-sense (5´-UGAUACUGCGCAAUUGCUGdTdT), designed against AcPRMT5, were used. The siRNA (4 μg) was transfected into A. castellanii trophozoites at a cell density of 4×105 per well, as previously described [17].
Statistical analysis
Data are expressed as mean±SEM. Statistical significance was analyzed using an unpaired Student’s t-test. A P-value of <0.01 was interpreted as statistically significant.
RESULTS
Identification of protein arginine methyltransferase 5
To identify the encystation-mediating factor of A. castellanii, we used the basic local alignment search tool (BLAST) to screen the cDNA library and identified a full-length open reading frame of the protein arginine methyltransferase 5 (AcPRMT5) (GenBank no. KT345169). AcPRMT5 contains domains found in S-adenosylmethionine-dependent methyltransferases (79-148 amino acids) and in PRMT5 arginine-N-methyltransferase (29-281 amino acids). Amino acid sequence alignment of AcPRMT5 with the PRMT5 of Plasmodium reichenowi, Plasmodium falciparum, Dictyostelium discoideum, Entamoeba histolytica, Trichomonas vaginalis, and Trypanosoma brucei showed sequence similarity in the methyltransferase motifs I, post I, II, and III (Fig. 1).
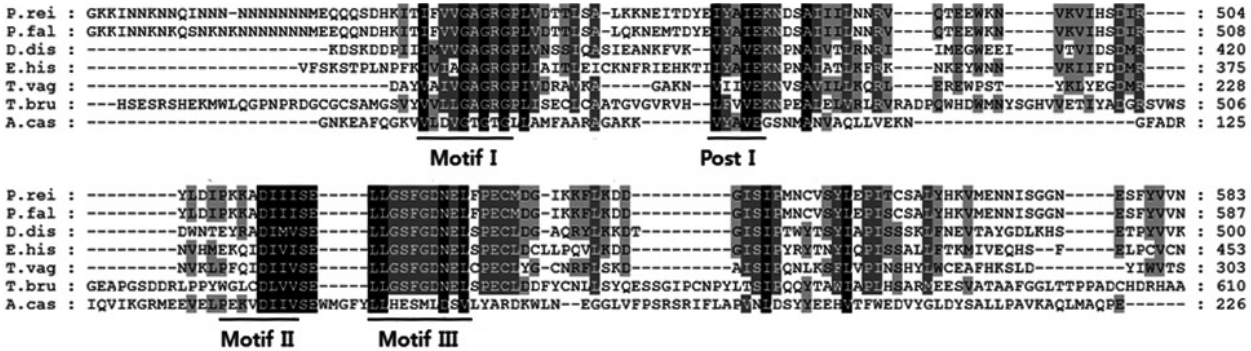
Amino acid sequence alignment of Acanthamoeba castellanii PRMT5 with PRMT5 of Plasmodium reichenowi, Plasmodium falciparum, Dictyostelium discoideum, Entamoeba histolytica, Trichomonas vaginalis, and Trypanosoma brucei. Sequences characteristic of methyltransferase motifs I, post I, II, and III are underlined.
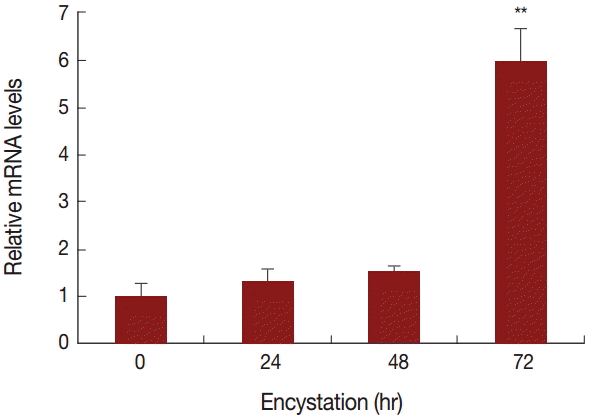
To compare the mRNA expression levels of AcPRMT5 during encystation, we conducted real time-PCR. The expression level of AcPRMT5 was highly increased 72 hr after the encystation induction (Fig. 2).
Intracellular localization of AcPRMT5 during encystation
A. castellanii EGFP-PRMT5 fusion protein showed a weak expression in the cytoplasm but a strong expression in the nucleus of trophozoites (Fig. 3A). During encystation, EGFP-PRMT5 fusion protein showed a similar localization pattern, with most of the protein localized in the nucleus of cysts (Fig. 3B). These results were confirmed using DAPI staining.
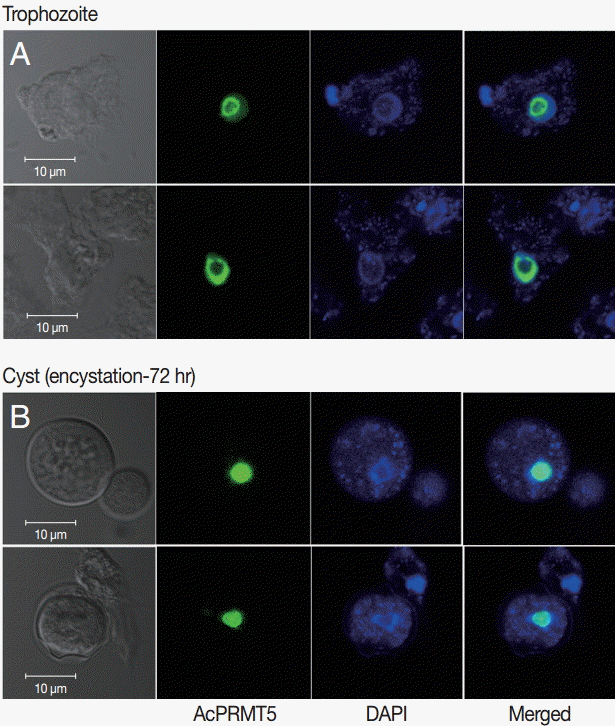
Stable transfection of pGAPDHgPRMT5 for intracellular localization. (A) EGFP-PRMT5 fusion protein of A. castellanii was mainly localized in the nucleus of trophozoites. (B) After encystation, EGFP-PRMT5 fusion protein was localized in the nucleus of cysts. Localization of AcPRMT5 in the nucleus was confirmed by DAPI staining.
Gene silencing of AcPRMT5 by siRNA
To determine the effect of PRMT5 during A. castellanii encystation, gene silencing by siRNA was performed. In the siRNA transfected A. castellanii, PRMT5 did not have a high expression level during encystation (Fig. 4A). Furthermore, after 72 hr of encystation induction, the formation of mature cysts was inhibited (20.4%) (Fig. 4B).
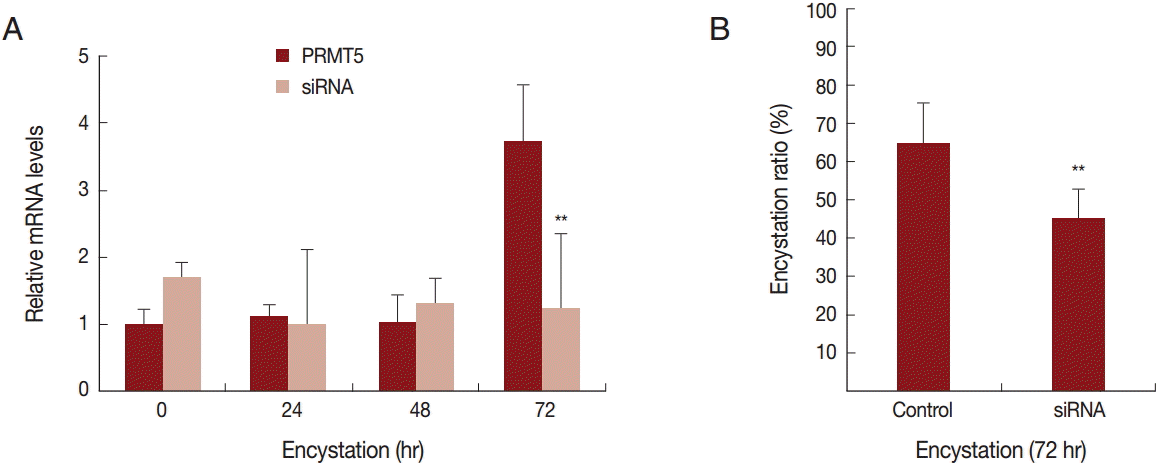
Transfection of A. castellanii with siRNA designed against PRMT5. (A) PRMT5 showed a high expression level during encystation ( ), but was down-regulated in PRMT5-siRNA transfected cells (
), but was down-regulated in PRMT5-siRNA transfected cells ( ). (B) This inhibition by PRMT5-siRNA was found to reduce encystation ratios. Values indicate the mean (±SD) of 3 experiments. **Means are significantly different one another (Student’s t-test, P<0.01).
). (B) This inhibition by PRMT5-siRNA was found to reduce encystation ratios. Values indicate the mean (±SD) of 3 experiments. **Means are significantly different one another (Student’s t-test, P<0.01).
DISCUSSION
Post-translational modification of proteins by arginine methylation is involved in a variety of cellular processes including transcriptional regulation [18]. Protein arginine methyltransferases (PRMTs) transfer methyl groups from S-adenosylmethionine (AdoMet) to arginine residues resulting in methylated arginines [19]. Three regions of sequence similarity, motif I, II, III, and post I have been reported in several AdoMet-dependent methyltransferases [20,21]. Methyltransferase motif I comprises 9 residues with the consensus sequence (V/I/L)(L/V)(D/E)(V/I)G(G/C)G(T/P)G, motif II comprises 8 residues with the consensus sequence (P/G)(Q/T)(F/Y/A)DA(I/V/Y)(F/I)(C/V/L), and motif III comprises 10 conserved residues LL(R/K)PGG(R/I/L)(L/I)(L/F/I/V)(I/L) after the end of motif II. The high degeneracy and narrow width of the post I motif did not allow its direct identification [20]. Post I was, therefore, appended to the description of motif I, considering 2 spacings: motif I-(10-30)-post I and motif I-(5-25)-post I. It was shown that post I comprised 5 residues with the consensus sequence (V/I/L/F/Y/M/H)(I/V/L/F/Y/T/K/Q/D)(G/A/S/I/D)(V/I/L/N/S/F/C/A)(D/E/N) [20]. The AcPRMT5 identified in this study showed high similarity to motif I, post I, and motif II (Fig. 1). Motif I is involved in AdoMet binding [20].
Many known arginine methyltransferases are proteins that have the ability to shuttle between the nucleus and the cytoplasm [22,23]. In mammalian cells, PRMT5 is localized in both the cytoplasm and the nucleus, where it is involved in the methylation of multiple histone and non-histone proteins [24]. Most of the AcPRMT5 was localized in the nucleus of trophozoites and cysts, while not much was found in the cytoplasm (Fig. 3). We hypothesize that AcPRMT5 regulates the transcriptional levels of encystation-associated genes by histone modification in the nucleus.
AdoMet-dependent PRMTs are generally classified into type I and type II enzymes. PRMT1 is the predominant member of type I PRMTs, whereas PRMT5 is a representative member of type II PRMTs. PRMT1-mediated dimethylation of arginine 3 of histone 4 (H4R3) is associated with gene activation [25], while the methylation of H4R3 by PRMT5 is associated with gene repression [26]. The target substrate of AcPRMT5 has not been identified yet. However, the inhibition of encystation by AcPRMT5-siRNA transfection (Fig. 4) suggests that AcPRMT5 regulates encystation-mediating factors in A. castellanii. To our knowledge, no PRMT substrates have been identified in Entamoeba spp., nor have any functional studies been performed. Recently, PRMT1a of E. histolytica was reported to be associated with gene expression regulation by dimethylation of H4R8 [27]. T. gondii CARM1 (PRMT homologue) resulted in decreased parasite viability due to the transient expression of enzymedead mutant TgCARM1 in tachyzoites [28]. RNAi-mediated knockdown of T. brucei PRMT1 resulted in a slow growth phenotype and abnormal cell cycle progression [29]. In rat liver cells, PRMT5 negatively affected cyclin E1 promoter activity and cellular proliferation [30]. It was suggested that cyclin E1 is a key mediator of PRMT5-dependent cell cycle arrest [30]. In A. castellanii, we consider that PRMT5 is associated with cell cycle arrest. AcPRMT5 was highly expressed at a late stage of encystation (Fig. 2), and the inhibition of AcPRMT5 reduced the encystation ratio (Fig. 4B).
In this study, we described for the first time the role of protein arginine methyltransferase 5 as a regulator of encystation mediating factors in Acanthamoeba. Epigenetic regulators play an important role in control of gene expression by modifying the chromatin. Epigenetic factors are, therefore, important for development of many diseases. Further efforts should be made to understand how the epigenetic mechanisms of parasites may be used in therapeutic treatments.
Acknowledgements
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2014R1A1A 2058405).
Notes
We have no conflict of interest related to this work.