Isolation and Genotyping of Acanthamoeba spp. as Neglected Parasites in North of Iran
Article information
Abstract
Acanthamoeba, a free-living amoeba, is widely distributed in the environment, water sources, soil, dust, and air. It can cause keratitis in contact lens wearers with poor hygiene and also fatal granulomatous amebic encephalitis (GAE) in immunocompromised hosts. The aim of this study was to gain some insights into the distribution and genotypes of the potentially pathogenic species of Acanthamoeba present in water sources in north of Iran. Total 43 Acanthamoeba species were isolated from 77 water samples taken from different water sources within the Mazandaran province in Northern Iran (Sari city and suburbs). Isolates were identified based on cyst and trophozoite morphological characteristics as well genetics. PCR fragments corresponding to the small-subunit 18S rRNA gene were sequenced for 20 of 43 positive isolates. The results revealed that 83.3% of sequenced isolates belonged to the T4 genotype and the rest belonged to the T2 genotype. Our results indicated that Acanthamoeba is widely distributed in Sari city. As the incidence in Iran of amoebic keratitis has increased in recent years, the exact estimation of the prevalence of this amoeba and its predominant genotype may play a crucial role in prevention of the disease. Sari city has several rivers, seashores, and natural recreational amenities, which attract visitors during the year. This is the first report of Acanthamoeba genotypes from water sources in Sari city, Mazandaran province of Iran, and the results suggest that more attention is needed to protect the visiting population and immunocompromised individuals.
INTRODUCTION
Acanthamoeba species are the most ubiquitous free-living amoeba (FLA) found in environments, including soil, air, and water [1,2]. Acanthamoeba has been isolated from water supplies, ocean, freshwater, seawater, tap water, hot tubes, irrigation water, bottle water, aquariums, plants, swage, sludge, dental unites, contact lens cases and solutions, air dusts, and even in animal feces [3,4]. It has been shown in some studies that Acanthamoeba can be resistant to chlorination but this is not common worldwide [5,6].
There are 2 stages in the life cycle of Acanthamoeba spp.: a vegetative trophozoite stage and a resistant cyst stage. Trophozoites are about 12-23 μm in diameter but it varies between different genotypes and species. During the trophozoite stage, Acanthamoeba feeds on bacteria, yeast, algae, human cells, and other microorganisms. The cyst stage is 5-20 μm in diameter, but, again, this varies according to genotype. Cysts are airborne and can spread easily in the environment and also to susceptible hosts [7].
Acanthamoeba can cause fatal granulomatous amoebic encephalitis (GAE), eye keratitis [8] skin ulceration and sometimes nasopharynx involvement [9,10]. Of 17 genotypes, the genotype T4 is the most frequently involved in cases of Acanthamoeba keratitis (AK) and GAE all around the world [3,11,12].
Rezaaeian and Niyyati reported the first case of AK in 1994 in a contact lens wearer with inadequate lens care. It is believed that AK has increased during the last decade in Iran, as well as the number of immune-deficient cases at risk of catching GAE [13]. To make matters worse, Acanthamoeba acts as a Trojan horse and carries microorganisms such as Mycobacterium avium, Vibrio cholerae, Listeria monocytogenes, Legionella pneumophila, and Burkholderia cepacia, which can then cause infection in the host [2,14,15]. Due to all these threats and their potential impact on human health, early detection of pathogenic Acanthamoeba in aquatic environments is crucial.
Sari is located in Mazandaran, a northern province of Iran. The proximity of the city to the Caspian Sea, as well as the presence of several rivers and natural recreational amenities, attracts visitors from all over Iran. As such, every year, many people visit the city and swim in the rivers, the natural lakes, and the sea. Until now, there the prevalence of Acanthamoeba in this province remained unknown. Therefore, the aim of this study was to gain some insights into the distribution and genotypes of the potentially pathogenic species of Acanthamoeba present in the water sources of Sari. In fact, we believe that this is the first molecular and morphological characterization of Acanthamoeba from water sources in Mazandaran province of Northern Iran.
MATERIALS AND METHODS
Sampling area
This study was carried out in Sari city, and its close suburbs, located in the Mazandaran province in Northern Iran, close to the Caspian Sea from May-June 2013. Mazandaran is a province with 22 townships with humid climate and 700 mm of rainfall annually. Sari is the capital of the province and has plenty of water sources. Therefore, water samples were taken from lakes, rivers, waterscapes, sea, tap waters, pools, waterholes, rice fields, and fishponds. Briefly, 77 samples were collected in duplicate from each water source located in either Sari or its suburbs. After collection, 250 ml sterile bottles containing the samples were transferred immediately to the department of parasitology and mycology of Mazandaran University of Medical Science for further analysis.
Isolation and culture of trophozoites
Samples were filtered through a cellulose nitrate filter (Millipore Corporation, Bedford, Madison, USA), pore size 0.45 μm. In order to isolate Acanthamoeba, the filters were inverted onto 1.5% non-nutrient agar plates coated with heat-killed Escherichia coli. The plates were sealed with paraffin film and incubated at 37˚C for up to 2 months. The presence of cysts and trophozoites was controlled daily for 1 month using an inverted microscope.
Morphological analysis
Culture plates containing Acanthamoeba cysts and trophozoites were observed microscopically, and characterization was performed according to the Pussard and Pons criteria [16]. Plates were stored for up to 2 months, and positive plates (plates exhibiting amoeba growth) were subjected to the following processes.
DNA extraction
Positive plates were gently scraped and washed with PBS (pH 7.4) 3X at 2,000 rpm for 5 min. After resuspending the cell pellets in lysis buffer (50 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0), they were incubated with proteinase K (0.25 mg/ml) at 56˚C overnight. Proteinase K was then deactivated by putting the samples into boiling water for 20 min. Finally, DNA was extracted using a phenol-chloroform method [17].
PCR assay
Genus specific primers for Acanthamoeba spp. were used to amplify the DF3 region [18], 423-551 bp, of the 18S rRNA (JDP1:5ʹ-GGCCCAGATCGTTTACCGTGAA-3ʹ) and (JDP2: 5ʹ-TCTCACAAGCTGCTAGGGGAGTCA-3ʹ). PCR was performed in a PCR thermoycler (BIO-RAD, Hercules, California, USA) in 20 μl volume of PCR pre-mix (Bioneer, Seoul, Korea), 1 μl of 10 pmol of each primer, and 4 μl of genomic template DNA. PCR products were visualized on 1.5% agarose gel containing ethidium bromide.
Nucleotide sequencing
For genotype identification, partial 18S rRNA sequencing (DF3 region) was performed with amplification primers and/or appropriate sequencing primers. Sequencing was performed for 20 of 43 positive samples and compared with reference species for each genotype.
Maximum-likelihood reconstructions were performed using the phylogenetic program MEGA version 6 using the GTR model of evolution G with 6 rate categories across sites, with 2,000 bootstrap replicates. The pairwise distance (Tr + Tv) was used. Nucleotide differences between the genotypes and clusters were calculated on the basis of the total 18-rRNA with the MEGA program. The calculation was based on the p-distance with pairwise deletions and without selected sites.
RESULTS
Our aim of this study was isolation and identification of Acanthamoeba spp. from Sari city and its suburbs (Mazandaran province) (Fig. 1). Forty-three (55.8%) Acanthamoeba spp. were isolated from 77 samples from several parts of Sari city and its suburbs according to morphological criteria of both cysts and trophozoites (Fig. 2) and molecular methods (PCR products, 423-550 bp) (Fig. 3).
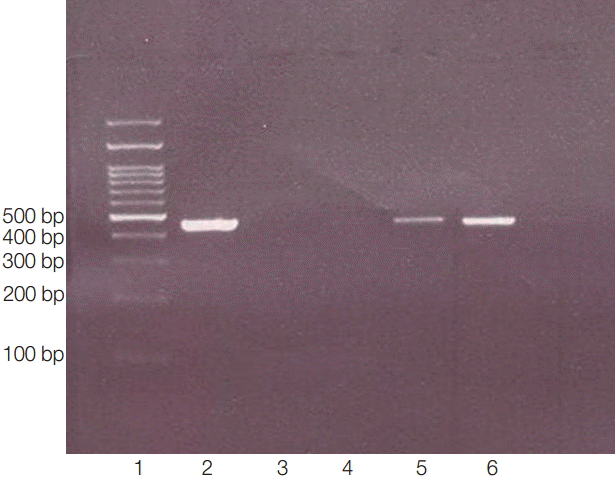
PCR products of 18s rRNA made visible bands in 423-550 bp. Lane 1, ladder 100 bp; lane 2, positive control (obtained from previously sequenced sample); lane 3: blank control (without DNA); lane 4, negative control (genomic DNA obtained from Cryptococcus neoformance); lanes 5-6, positive isolates.
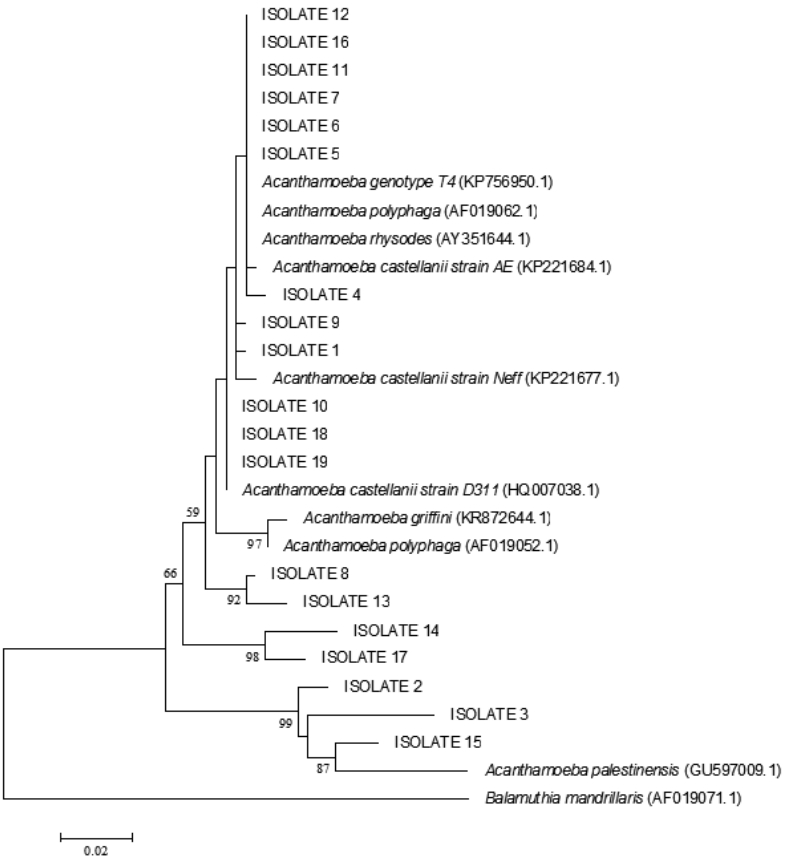
Twenty positive isolates were sent for sequencing, among which, 2 sequences were not perfectly recognized and thus omitted from the study. Sequencing results showed that 83.3% (15 samples) of all positive samples belonged to Acanthamoeba T4 genotype, while 16.7% (3 samples) belonged to A. palestinensis T2 genotype (Table 1). Sequence information obtained in this study was deposited in GenBank, and accession numbers for our nucleotide sequences were received as KU872061, KU872062, KU872063, and so on.
In blast search and analysis of 18 sequences, 15 isolates from rivers, canals, ditches, pipes, rice fields, and sea sediments showed high homology with Acanthamoeba genotype T4. Among these 15 isolates belonging to the Acanthamoeba genotype T4, 4 were defined as A. rhysodes, 2 as A. polyphaga, and 9 as A. castellanii. As mentioned earlier, 3 isolates from rice field and fish breeding ponds were identified as A. palestinensis T2 genotype. The phylogenetic tree showed correlation between isolates and Acanthamoeba spp. (Fig. 4).
DISCUSSION
Acanthamoeba is an amphizoic opportunistic protozoan parasite that is widely distributed in the environment as well as in tap water, contact lens cases, soil, dust, and air and is known to produce serious human infections [19]. Acanthamoeba species can cause either chronic granulomatous encephalitis or chronic amoebic keratitis as they can carry potential dangerous bacteria such as Legionella, Mycobacterium, and Pseudomonas [20,21]. The existence of Acanthamoeba in different environmental and water sources worldwide leads us to analyze water sources from Mazandaran province in Iran.
Mazandaran is a vast province in northern Iran, and annually many visitors from all parts of Iran travel to Sari city to enjoy the beach and to swim in the sea. Its many water sources, such as lakes and rivers, make this province a place for recreation, and therefore, water contact is inevitable. To our knowledge, this is the first study in Mazandaran province to document Acanthamoeba spp. Our research performed on water sources used in Mazandaran province (Sari city and its suburbs) revealed evidence for the presence of Acanthamoeba spp. in human habitation. Acanthamoeba is a potential pathogen especially for immunocompromised individuals and it is also a risk factor for contact lens wearers and even for non-contact lens wearers [4,7].
Several studies have been carried out in water sources from Iran. Maghsood et al. [9] examined 12 pool and waterfall samples, from which Acanthamoeba T4 was found to exist in 58.3% of the pool samples and 33.3% of the waterfall samples [9]. In our study, 83.3% of positive samples gathered from different water sources along the city belonged to the Acanthamoeba T4 genotype. Niyyati et al. [18] isolated Acanthamoeba from 50 keratitis samples and 10 environmental samples, from which 86.7% of positive samples belonged to the T4 genotype. In addition, they identified T3 and T11 genotypes [18]. These results are similar to our findings although, in our study, we isolated 3 A. palestinensis belonging to the T2 genotype, while the rest of the Acanthamoeba isolates were classified as belonging to the T4 genotype.
The Acanthamoeba genotype most related to keratitis is T4 [3,9,22,23]. Free-living amoebas, including the Acanthamoeba T4 corresponding to A. castellanii, were isolated from hot spring water in Ardebil city by Badirzadeh et al. [24]. We also isolated A. castellanii from the river, rice field, and pound water. These findings were particularly relevant, being that some of the travelers visiting Mazandaran province like to swim in the river and so the amoeba can be potentially dangerous for them. In addition, according to the results from a study carried out by Rezaeian et al. [25], Acanthamoeba spp. were isolated from 46.3% of their samples obtained from numerous localities [25]. Moreover, in another study carried out by Bagheri et al. [26], Acanthamoeba was isolated from 48% of the tap water of 13 hospitals located in different parts of Iran.
A study by Nazar et al. [17] conducted on 50 parks and squares from Tehran found that 32% of their collected samples were positive for Acanthamoeba spp. and that 87.5% of these positive isolates belonged to the T4 genotype and 12.5% to the T5 genotype. In addition, 17% of our isolated amoeba were defined as A. palestinensis T2 genotype and were isolated from the rice field. This finding is especially important for farmers working in these farms as this genotype had previously been reported as a causative agent of amoeba-related keratitis [27]. Reports from other parts of Iran showed similar results. Rahdar et al. [28] isolated Acanthamoeba spp. in 71.6% of water samples and 26% of soil samples collected in Ahvaz city, among which 86.6% belonged to T4 genotype, 6.6% to T2 and 6.6% to T5.
In our study, the T4 genotype was the most abundant one, having been recognized in 83.3% of samples used in genotyping, which makes it an important risk for human health in this geographical region. This result is in accordance with previous findings that suggested T4 as the most prevalent genotype in the world [18,29]. In conclusion, the incidence of AK in Iran is increasing due to an increase in contact lens wearing and poor hygiene behaviors among users. Moreover, immunocompromised and HIV+ populations are at risk with this amoeba existing in their water sources. Therefore, it is important to develop programs to promote awareness towards the existence of the potential pathogenic amoeba and thus better hygienic practices among contact lens wearers and visitors to Sari city. In addition, it may be useful to titer antibodies in HIV+ and immunocompromised residents in hospitals to prevent unsafe water contact. These results should be made available for all physicians in order to manage the patients and susceptible populations with adequate care and to organize proper control programs.
Acknowledgements
The authors would like to appreciate very much for kind collaboration of Mr. Azami. They also would like to thank the financial support by Vice Chancellors for Research of Mazandaran University of Medical Sciences, Iran (project no. 92-28).
Notes
The authors declare that they have no competing interests.