Subtype Distribution of Blastocystis in Communities along the Chao Phraya River, Thailand
Article information
Abstract
Blastocystis is a common zoonotic enteric protozoan that has been classified into 17 distinct subtypes (STs). A cross-sectional study was conducted to determine the prevalence and subtype distributions of Blastocystis in villagers living along the Chao Phraya River, Ayutthaya Province, Thailand, and to assess the risk of zoonotic infection. In total, 220 stool samples were collected, and DNA was extracted. PCR and sequencing were performed with primers targeting the small-subunit ribosomal RNA (SSU rRNA) genes. Blastocystis was present in 5.9% (13/220) of samples, and ST3 (5.0%; 11/220) was the predominant subtype, followed by ST2 (0.45%; 1/220) and ST6 (0.45%; 1/220). Phylogenetic trees were constructed with the maximum-likelihood method based on the Hasegawa–Kishino–Yano + G + I model, neighbor-joining, and maximum parsimony methods. The percentage of bootstrapped trees in which the associated taxa clustered together was relatively high. All the sequences of the Blastocystis-positive samples (KU051524–KU051536) were closely related to those from animals (pig, cattle, and chicken), indicating a zoonotic risk. Therefore, the villagers require proper health education, especially regarding the prevention of parasitic infection, to improve their personal hygiene and community health. Further studies are required to investigate the Blastocystis STs in the animals living in these villages.
INTRODUCTION
Blastocystis is an enteric zoonotic protozoan found in humans and many kinds of animals [1]. It is one of the most commonly detected protozoans in parasitological surveys, with high infection rates in developing countries [2]. Transmission can occur from humans to humans, humans to animals, and animals to humans via the fecal-oral route.
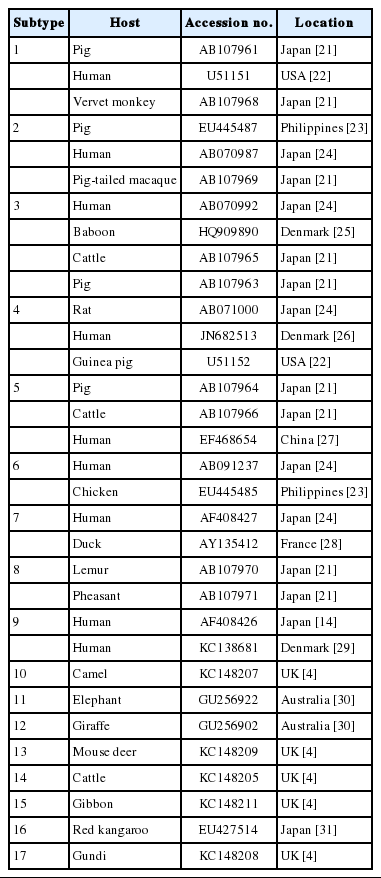
Several methods are used to diagnose Blastocystis infection, including direct examination, culture, and PCR. Among these, PCR is the most powerful, sensitive, and specific detection technique for Blastocystis and can be used for subtyping analyses [3]. At least 17 distinct subtypes (STs) of Blastocystis have been identified in humans, non-human primates, mammals, and avian hosts, based on the analysis of the small-subunit ribosomal RNA (SSU rRNA) genes [4]. ST1-ST9 have been reported in humans, and ST1-ST4 are most common [5,6]. Some Blastocystis STs detected in humans have also been identified in animals. This suggests that animals can act as reservoirs for this protozoan and may be linked to its zoonotic transmission. Therefore, subtyping of this protozoan allows us to understand the interactions between humans and animals in specific communities.
The Chao Phraya River is one of the most important water sources in Thailand. Many communities and animal husbandry activities are located along this river. Importantly, it is the source of water for the people living along it, including those in Ayutthaya Province. It is possible that domestic animals, wildlife, and communities shed intestinal parasites into the river, readily spreading them to the environment, and thus distributing parasitic diseases among the communities.
Until now, limited data have been available on the prevalence and subtypes of Blastocystis in the communities living along the Chao Phraya River. Such data could be used to support and promote the health and quality of life of the humans and animals in the area, and to improve the interactions between them. In this study, we investigated the prevalence and subtype distributions of Blastocystis among the villagers living along the Chao Phraya River.
MATERIALS AND METHODS
Collection of human stool samples
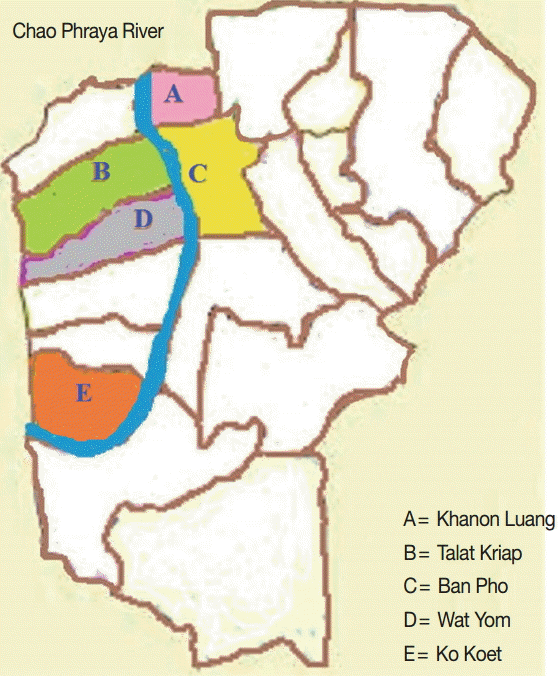
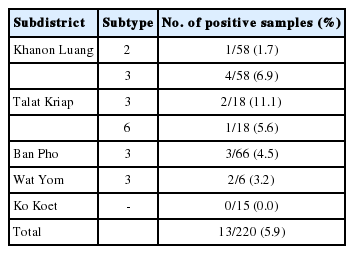
A cross-sectional study was conducted in September-October 2015 in 5 subdistricts (Khanon Luang, Talat Kriap, Ban Pho, Wat Yom, and Ko Koet) in the Bang Pa-In district of Ayutthaya Province, in central Thailand. These subdistricts are situated along the sides of the Chao Phraya River, and the villagers work in agriculture and live in close contact with animals, as shown in Fig. 1. Moreover, there has been some concern about the health of the villagers. In total, 220 human stool samples were collected. The participants ranged in age from 1 to 75 years. No participant complained of any gastrointestinal symptoms, such as abdominal pain or diarrhea. The specific instructions for collecting the stool samples and avoiding their contamination were clearly explained to all the participants. The stool samples were kept cool during transportation to the laboratory, and were then stored at -20˚C until DNA extraction. The study protocol was approved by the Ethics Review Committee for Research Involving Human Research Subjects, Health Sciences Group, Chulalongkorn University, Bangkok, Thailand (COA no. 152/2558).
DNA extraction from stool samples and PCR amplification
DNA was extracted directly from all the fresh stool samples using the PSP® Spin Stool DNA Kit (STRATEC Molecular GmbH, Berlin, Germany), according to the manufacturer’s instructions. DNA samples were stored at -20˚C until analysis.
Blastocystis was detected, and the STs were characterized with a nested PCR analysis of the DNA from the collected stool samples. The primer sets and conditions used have been published previously and are specific for the partial SSU rRNA genes, which differentiate the subtypes of Blastocystis: external primer set, RD3 5ʹ-GGGATCCTGATCCTTCCGCAGGTTCACCTAC-3ʹ and RD5 5ʹ-GGAAGCTTATCTGGTTGATCCTGCCAGTA-3ʹ[7]; and internal primer set, forward 5ʹ-GGAGGTAGTGACAATAAATC-3ʹ and reverse 5ʹ-ACTAGGAATTCCTCGTTC ATG-3ʹ [2]. PCR products were about 1,100 base pairs (bp) in length and were detected with 1.5% agarose gel electrophoresis. DNA in the gels was stained with ethidium bromide and visualized under a transilluminator.
DNA sequencing and phylogenetic reconstruction
All the positive samples were sequenced with an ABI 3730 XL sequencer, using fluorescent-dye terminator sequencing by Bio Basic Canada Inc. (Ontario, Canada). The positive sample sequences were read and edited with BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The Blastocystis STs of the positive samples were identified by comparing them with the 17 Blastocystis STs available in the GenBank database using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) as shown in Table 1. A multiple sequence alignment was constructed with Clustal W [8]. Phylogenetic reconstructions were performed with MEGA 6.0 [9]. The phylogenetic trees were constructed with maximum-likelihood (ML) method based on the Hasegawa-Kishino-Yano + G (gamma distribution) + I (invariant) model (best-fit substitution model), and also neighbor-joining (NJ) and maximum parsimony (MP) methods. Bootstrap values were calculated with 1,000 replicates. Proteromonas lacertae (accession no. U37108), which is closely related to Blastocystis, was used as the outgroup.
Statistical analysis
A descriptive analysis of percentages was used to explore the positive stool samples and ST distributions of Blastocystis.
RESULTS
Prevalence of Blastocystis determined with nested PCR
The overall prevalence of Blastocystis infection detected with nested PCR in this study was 5.9% (13/220). Fig. 2 shows positive PCR results for Blastocystis. PCR products were about 1,100 bp in length.
Subtype characterization of Blastocystis in positive stool samples
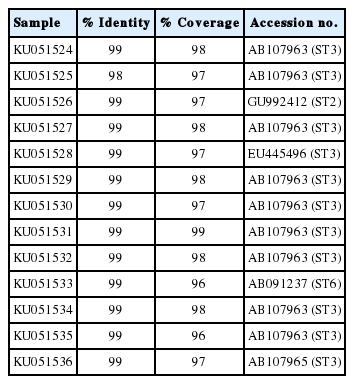
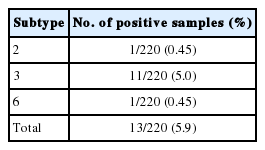
The Blastocystis STs were identified with direct DNA sequencing. BLAST results for positive samples were shown in Table 2. Only ST3, ST2, and ST6 were detected. ST3 (5.0%; 11/220) was the predominant subtype, followed by ST2 (0.45%; 1/220) and ST6 (0.45%; 1/220) as shown in Table 3. Table 4 shows the ST distributions of Blastocystis according to geographic subdistricts. The ages of the Blastocystis-infected individuals ranged from approximately 30 to 70 years, except for 2 infected children from Talat Kriap and Khanon Luang, whose ages were 1 year (ST6 infected) and 8 years (ST3 infected), respectively.
Phylogenetic reconstruction and genetic divergence of Blastocystis isolates
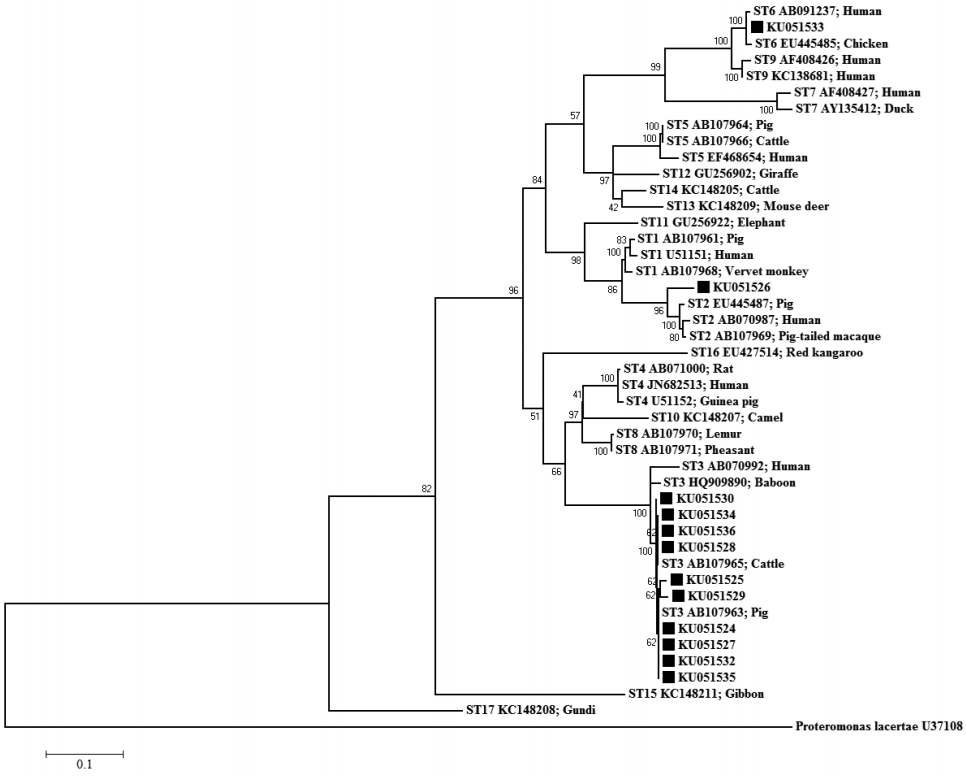
The ML tree of Blastocystis constructed from SSU rRNA gene sequences is shown in Fig 3. The ML method was based on the Hasegawa-Kishino-Yano+G+I model. The percentage of bootstrapped trees in which the associated taxa clustered together was relatively high. The ML tree, upon which the SSU rRNA gene sequences of the Blastocystis-positive samples were compared with 17 reference sequences in the GenBank database, classified KU051524, KU051525, KU051527-KU051530, KU051532, and KU051534-KU051536 as ST3, KU051526 as ST2, and KU051533 as ST6. All the sequences from the Blastocystis-positive samples were closely related to animal sequences (from pigs, cattle, and chickens). The NJ and MP trees showed the same clustering patterns as the ML tree (data not shown).
DISCUSSION
The relatively low (5.9%) prevalence of Blastocystis observed in this study might be attributable to the characteristics of the study area, where public utilities, such as tap water, are available to almost every household. Therefore, the villagers are protected from infection because they use treated water in their daily activities. The prevalence of Blastocystis infection is higher in developing countries than in developed countries. However, it varies from country to country, and within the same country [2,10,11]. In Thailand, Blastocystis infection has been reported at rates as high as 37.2% [12]. Several risk factors seem to be associated with Blastocystis infection, including poor personal hygiene and high-risk behavior, poor community sanitation, socioeconomic status, lifestyle, and the culture of the population. Blastocystis can also infect a range of animals, including pigs, cattle, monkeys, and chickens. Zoonotic STs have been isolated from these animals, and they may therefore act as reservoir hosts for this protozoan and facilitate its zoonotic transmission. Zoonotic transmission is considered an important mode of Blastocystis infection in humans, and most infections occur via the fecal-oral route.
To date, the genetic diversity of this protozoan has been demonstrated with SSU rRNA gene analyses, which have detected at least 17 distinct STs [4]. ST1-ST9 have been reported in humans, and ST3 is the most prevalent. In this study, ST3 was the predominant subtype, followed by ST2 and ST6. The findings of this study are consistent with those of several studies conducted in many countries, including Singapore, Turkey, Bangladesh, and Egypt [2,6,13,14], which all reported ST3 as the predominant subtype. However, this subtype has also been isolated from animals such as pigs, dogs, cattle, and non-human primates [15]. Surprisingly, ST1 was not detected in the present study, although several studies in Thailand have reported that ST1 is a common subtype [16-18]. This discrepancy could be attributable to differences in the study areas and study populations investigated, revealing different subtype distributions. We also observed ST6 in a 1-year-old child. ST6 is considered a typical ‘avian’ subtype [19], and is rare in nonavian hosts. The infection may have been transmitted by the unwashed hands of the parents or carers, after their contact with animals, or when the child was crawling on the ground in areas contaminated with Blastocystis cysts. This clearly demonstrates that animals are potential risk factors for Blastocystis infection and should be considered zoonotic sources.
In this study, the PCR primers that we used amplified the partial SSU rRNA gene [2], which contains highly variable nucleotides that allow Blastocystis to be differentiated into subtypes. This sequence also allows the construction of reliable phylogenetic trees. We believed that it is one of the best techniques for Blastocystis subtyping, instead of sequencing complete SSU rRNA gene. However, the ‘barcode region’, a 600-bp 5ʹ region of Blastocystis SSU rRNA, is also recommended for Blastocystis subtyping [20]. It reduces the time and the costs of the procedure insofar as the sequence can be amplified with only a single round of PCR. Therefore, the ‘barcode’ PCR is a useful alternative method for large epidemiological studies.
The ML tree indicated that all the Blastocystis-positive samples were potentially involved in zoonotic transmission, as shown in Fig. 3. ST3 has been reported in pigs, but is uncommon. Thathaisong et al. [18] demonstrated that ST1 was common in pigs kept at an army base in Chonburi Province. In the present study, the individuals infected with ST3 may have acquired Blastocystis from pigs (person-to-pig or pig-to-person transmission via the fecal-oral route), because we observed that their livestock (pigs, chicken, and cattle) were regularly kept under or near their houses. However, stools must be collected from all the animals in the infected houses to test this assumption. It is difficult to avoid the aerosols emitted from animal stools, which could contribute to the transmission of Blastocystis in these houses (on the ground), and frequent animal contact may increase the risk of Blastocystis infection in this area.
In conclusion, the risk of Blastocystis zoonotic infection in these villagers was considered in this study. This risk can involve a combination of factors, including the traditional culture, human activities, and animal contact (pigs, cattle, and chickens), all of which increase the risk of Blastocystis infection in the study area, even though the rate of Blastocystis infection was relatively low in this study. The villagers must have access to proper health education regarding the prevention of parasitic infections because they do not understand the dangers associated with contact with animals infected with pathogenic organisms. They should also be encouraged to improve their personal hygiene and community health. Further studies are required to investigate the Blastocystis STs in the animals living in these villages.
Acknowledgements
We would like to thank all the participants and staff of the Department of Clinical Microscopy, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, Thailand for their help in the collection of the stool samples. This research was supported by the National Research University Project, Office of Higher Education Commission, Thailand (WCU-58-001-HR).
Notes
The authors declare that they have no conflicts of interest.