Characterization of Caveola-Vesicle Complexes (CVCs) Protein, PHIST/CVC-8195 in Plasmodium vivax
Article information
Abstract
Plasmodium vivax produces numerous caveola-vesicle complex (CVC) structures beneath the membrane of infected erythrocytes. Recently, a member helical interspersed subtelomeric (PHIST) superfamily protein, PcyPHIST/CVC-8195, was identified as CVCs-associated protein in Plasmodium cynomolgi and essential for survival of this parasite. Very little information has been documented to date about PHIST/CVC-8195 protein in P. vivax. In this study, the recombinant PvPHIST/CVC-8195 N and C termini were expressed, and immunoreactivity was assessed using confirmed vivax malaria patients sera by protein microarray. The subcellular localization of PvPHIST/CVC-8195 N and C termini in blood stage parasites was also determined. The antigenicity of recombinant PvPHIST/CVC-8195 N and C terminal proteins were analyzed by using serum samples from the Republic of Korea. The results showed that immunoreactivities to these proteins had 61% and 43% sensitivity and 96.9% and 93.8% specificity, respectively. The N terminal of PvPHIST/CVC-8195 which contains transmembrane domain and export motif (PEXEL; RxLxE/Q/D) produced CVCs location throughout the erythrocytic-stage parasites. However, no fluorescence was detected with antibodies against C terminal fragment of PvPHIST/CVC-8195. These results suggest that the PvPHIST/CVC-8195 is localized on the CVCs and may be immunogenic in natural infection of P. vivax.
INTRODUCTION
Malaria is one of the most important infectious diseases of humans, causing almost 1 million deaths per year and extensive morbidity [1]. The 5 Plasmodium species known to cause malaria in human beings are P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi [2]. Among malaria parasite lifecycles, the erythrocytic stage is responsible for almost all clinical symptoms as well as associated morbidity and mortality of malaria [3]. Dramatic structural and morphological changes of parasites have been observed in erythrocytes after invasion by the parasite [4].
During the development of P. falciparum parasites inside red blood cells (RBCs), small protrusions called knob structures appear on the RBC surface, which play a major role in the pathophysiology of cerebral malaria [5–7]. Knob proteins play an essential role in malaria control since immunotherapeutic agents can be investigated to interrupt as well as reverse the mediation of cytoadherence [8]. The surface proteins are also potential vaccine candidates in spite of high antigenic variations [3,9]. In contrast to developing protruding knob structures in P. falciparum infected RBCs, P. vivax and simian vivax-type malaria species, such as P. cynomolgi, induce numerous small pink to red dots in Giemsa-stained blood films, known as Schüffner’s dots that are important in the identification of this species of malaria parasites and have been observed by electron microscopy as the caveola-vesicle complexes (CVCs) beneath the membrane of infected erythrocytes [10,11]. The CVC in P. vivax is composed of numerous flask-like indentations on infected reticulocytes membrane skeleton, called caveolae, associated with several tube-like vesicles [12–14]. The functional annotation of the CVCs remains either unknown or obscure. However, it has been suggested that CVCs may play an important role on nutrient uptake or release of metabolites from parasite-infected erythrocytes [10–13]. Immunoelectron microscopy of P. vivax-infected human erythrocytes using specific monoclonal antibodies confirmed the location of antigens within vesicles of CVCs [10,12,15]. Among those antigens, a 95 kDa parasite protein was shown to be a major parasite-derived component exclusively associated with CVCs [13]. Recently, this protein was recognized as a member of helical interspersed subtelomeric (PHIST) superfamily protein (PvPHIST/CVC-8195) [10]. The N-terminal of this protein contains export motif (PEXEL; RxLxE/Q/D) which has been shown to play an important role on trafficking of some proteins from the parasitophorous vacuole (PV) onwards to the RBC cytosol [16]. The protein’s PHIST domain, which contains 4 predicted alpha-helical domains and 4 positional conserved tryptophan residues, is located in the C-terminal region [17]. A further pcyphist/cvc-8195 gene disruption study indicated that this protein may be fundamental for the survival of malaria parasites [14]. To date, the antigenicity, as well as the localization of PvPHIST/CVC-8195 in P. vivax, have not been investigated.
In this study, 2 functional regions of PvPHIST/CVC-8195 N- and C-termini (NT and CT) were expressed. We sought to analyze the antigenicity of PvPHIST/CVC-8195 NT and CT using confirmed vivax patients sera by protein microarray and used immunofluorescence to confirm that PvPHIST/CVC-8195-NT is localized on the CVCs during the erythrocytic stage of P. vivax parasites.
MATERIALS AND METHODS
Human serum samples
Positive serum samples were collected from confirmed vivax patients by microscopic examinations at health centers and clinics in malaria-endemic areas of Gangwon Province, Republic of Korea (ROK). Their mean age was 24 years (range 18–52 years). Healthy control sera were collected from 32 malaria-naïve people living in non-endemic areas of Korea (no known malaria history). This study was approved by the Institutional Review Board at Kangwon National University Hospital.
Production of recombinant protein
Two fragments of pvphist/cvc-8195 (PlasmoDB no. PVX_093680) were amplified by PCR from cDNA as described previously [18]. Fragment PvPHIST/CVC-8195-NT comprised amino acid residues 1–170 of the full-length PvPHIST/CVC-8195 amino acid sequence (forward primer 5′-gatccccaggaattcccATGAGTCCCTGCAACATCC-3′ and reverse primer 5′-atgcggccgctcgagTTAAGCTGGTTGATCGGGCCTA-3′). Fragment PvPHIST/CVC-8195-CT comprised residues 556–710 (forward primer 5′-gatccccaggaattcccGACAATGAACAACTCCCATTCG-3′ and reverse primer 5′-atgcggccgctcgagTTAGAGTTTGCTGTGTTTCTTCATCT-3′). The underline and lowercase letter of primer sequence indicate homologous sequence to the vector sequence and restriction enzyme site, respectively. The PCR amplification products with high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) were ligated into the expression vector pGEX 4T-2 (GE Healthcare Life Sciences, Uppsala, Sweden). Positive clones were validated by DNA sequencing analysis. The correct clones were then transformed into competent BL21 StarTM (DE3) cells of E. coli (Invitrogen, Seoul, Korea) for protein expression. Soluble protein was purified on Glutathione SepharoseTM 4B (GE Healthcare Life Sciences) columns according to the manufacturer’s protocol.
SDS-PAGE and Western blot analysis
The PvPHIST/CVC-8195-NT and CT proteins were separated by SDS-PAGE under reducing conditions. Then, proteins were transferred from the SDS-PAGE gel to PVDF membranes (0.45 μm, Millipore, Billerica, Massachusetts, USA) by using a semi-dry blotting system (ATTO Corp., Tokyo, Japan) in a semi-dry transfer buffer (50 mM Tris, 190 mM glycine, 3.5 mM SDS, 20% methanol) at a constant 400 mA for 40 min and followed by blocking with 5% skim milk in PBS containing 0.2% Tween 20 (PBS-T) overnight. The mouse anti-penta His antibody (Qiagen, Hilden, Germany) and P. vivax or healthy control sera (1:200) were diluted into PBS/T and used as the first antibody. His-tagged recombinant proteins were finally detected by using goat anti-mouse (1:10,000, PBS/T) or goat anti-human (1:20,000, PBS/T) antibodies conjugated IRDye® (LI-COR Bioscience, Lincoln, Nebraska, USA). Reacted membrane was scanned by Odyssey infrared imaging system (LI-COR Biosciences) and results were analyzed by Odyssey software (LI-COR Biosciences).
Protein arrays
Amine-coated slides were prepared and used for protein microarray screening as described previously [19]. The humoral immune response was analyzed by using well-type amine arrays of sera from 100 patients with vivax malaria and 32 malaria-naïve individuals. Briefly, 1 μl of PvPHIST/CVC-8195-NT and CT (50 ng/μl) proteins, respectively, were loaded into duplicated array wells of individual sample, incubated at 37°C for 2 hr followed by blocking with 5% BSA in PBS-Tween (0.1%) (PBS-T) and then reacted with sera from malaria patients and healthy controls (1:10 dilution) in PBS/T for 1 hr at 37°C. Goat anti-human IgG conjugated Alexa Fluor 546 (10 ng/μL, Invitrogen) in PBS-T was used as the secondary antibody. The fluorescent signals were detected by fluorescence scanner (ScanArray Express, Perkin Elmer, Boston, Massachusetts, USA) and analyzed as described previously [20]. Two SDs above the mean fluorescence intensity (MFI) of the malaria-naïve samples were defined as the cutoff value. For normalization of the antibody array data, fluorescence intensities were divided by each cutoff value (Table 1).
Immunization of mice with PvPHIST/CVC-8195-NT and -CT
Six- to 8-week-old female BALB/c mice (DBL Co., Seoul, Korea) were injected intraperitoneally with ~20 μg of PvPHIST/CVC-8195-NT/CT and PBS as negative control with Freund’s complete adjuvant (Sigma-Aldrich, St. Louis, Missouri, USA). Three mice were used per group. Booster injections were given after 3 and 6 weeks using the same amount of antigen with Freund’s incomplete adjuvant (Sigma-Aldrich). Mouse blood samples were collected 2 weeks after the final boost.
Immunofluorescence assays (IFAs)
Intraerythrocytic stage P. vivax parasites were collected from a malaria patient in Korea. Parasite-infected blood smears were fixed with paraformaldehyde (4% in PBS) for 10 min at room temperature, and then dried and stored at −80°C. After thawing, slides dried in blue silica gel container (Samchun Chemical, Pyeongtaek, Korea) were blocked with nonfat milk (5% in PBS) at 37°C for 30 min. The slides were then incubated with 1:100 dilutions of mouse anti-PvPHIST/CVC-8195 immune sera followed by washing 3 times with cold PBS. The slides were stained with goat anti-mouse IgG conjugated with Alexa Fluor 488 or goat anti-rabbit IgG antibodies conjugated Alexa Fluor 568 (Invitrogen) and the nuclear stain 4′, 6-diamidino-2-phenylindole (DAPI; Invitrogen) at 37°C for 30 min, followed by washing 3 times with cold PBS. Finally, slides were mounted in Prolong Gold antifade reagent (Invitrogen) and fluorescence was visualized by using a confocal laser-scanning microscope (FV1000; Olympus, Tokyo, Japan). The FV10-ASW 3.0 Viewer software (Olympus) and Adobe Photoshop CS5 (Adobe Systems, San Jose, California, USA) software were used for capturing and preparing images for publication, respectively.
Data analysis
The TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used for searching transmembrane (TM) domains. The putative 3 dimensional structures were assessed using the Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/~phyre2/html/). Simple scatter-regression was used for making standard curve by SigmaPlot (Systat Software Inc., San Jose, California, USA). The GraphPad Prism software, version 5.0 (GraphPad, San Diego, California, USA) were used to determine the correlation between the duplicate spots of protein arrays as well as antibody reactivity of different concentrations of the recombinant proteins. Sensitivity was measured by percentage of patients who have fluorescence intensity higher than the cutoff value, and specificity was evaluated by percentage of healthy individuals who have fluorescence intensity lower than the cutoff value. The Student’s t-test was used to compare the differences between the means of each group for statistical significance. Statistical differences of P<0.05 were considered significant.
RESULTS
The complete PvPHIST/CVC-8195 protein sequence in the Sal-1 strain consists of 710 amino acids (a.a.) (molecular weight, 74.5 kDa). PvPHIST/CVC-8195 contains an ER signal peptide, a speculated plasmodium export element (PEXEL, RxLxE/Q/D), and 4 predicted alpha-helical domains (PHIST domain) (Fig. 1A). The 3-dimensional model of ER signal peptide in N-terminus and PHIST domain in C-terminus have been performed by phyre2 web portal [21] (Fig. 1B).
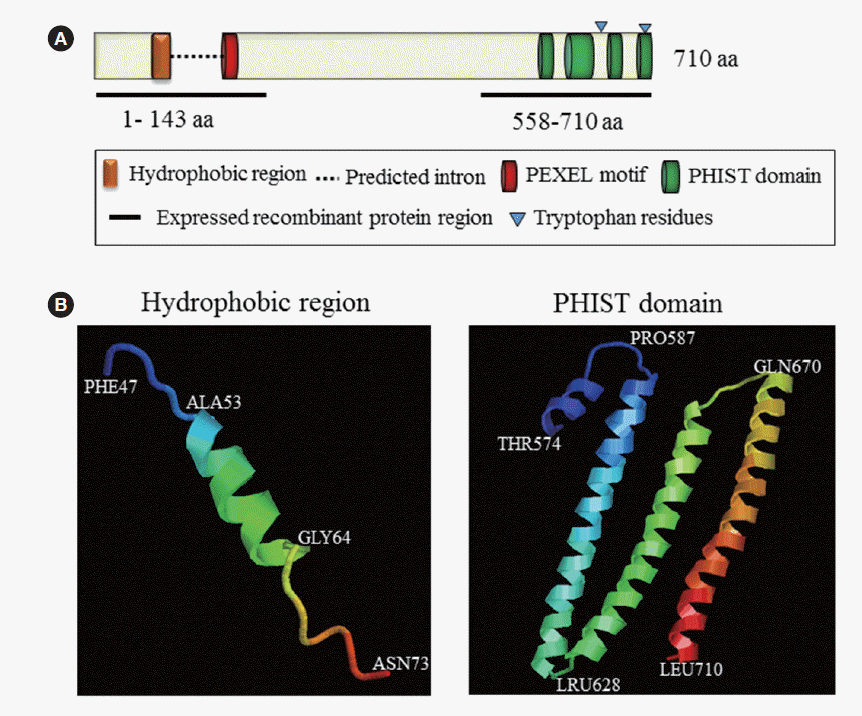
Schematic structure of PvPHIST/CVC-8195 and special domains. (A) The schematic diagram of PvPHIST/CVC-8195. (B) Predicted 3D structure of hydrophobic region and PHIST domain (http://www.sbg.bio.ic.ac.uk/~phyre2/html/).
In this study, the N-terminal (1–143 a.a.) and C-terminal (558–710 a.a.) fragments with GST-tag were designed and expressed using E. coli. The N and C terminal fragments of PvPHIST/CVC-8195, designated PvPHIST/CVC-8195-NT and PvPHIST/CVC-8195-CT, respectively, were produced as recombinant GST fusion proteins in E. coli. Western blot analysis using an anti-GST antibody, mouse immune sera, and vivax patients’ pooled serum detected PvPHIST/CVC-8195-NT and -CT as similar bands of ~45 kDa (Fig. 2A). The specific reaction of PvPHIST/CVC-8195 2 fragments with vivax patient serum samples suggested that the recombinant protein was expressed as least in part as a correctly folded form. Moreover, PvPHIST/CVC-8195 from the parasite antigen was recognized by immune sera as a single ~95 kDa band which was similar to PcyPHIST/CVC-8195 in P. cynomolgi (Fig. 2B) [14]. Pre-immune animal sera and non-infected human sera were also examined, but no band was detected (data not shown).

Recombinant protein expression and Western blot analysis of PvPHIST/CVC-8195. (A,B) Purified recombinant protein (rPro) of PvPHIST/CVC-8195-NT and -CT fragments were reacted with anti-GST antibody (GST), pre-immune mouse serum (PI), mouse immune serum (Mo), healthy individuals serum (H), and mixed vivax patients serum (Pt) samples. (C) Western blot analysis of erythrocyte lysate (RBC) P. vivax parasite lysate (RBC), parasite lysate (P) with anti-PvPHIST/CVC-8195-NT and -CT fragment immune serum samples.
To evaluate the humoral immune responses to PvPHIST/CVC-8195-NT and -CT fragments, a protein array of purified recombinant proteins were used to screen the presence of antibodies in human sera. Antibody responses against recombinant PvPHIST/CVC-8195 in serum samples from 100 vivax patients and 32 healthy individuals were determined.
An optimal concentration (50 ng/μl) of protein coating from amino-functionalized slide was determined using pooled sera from vivax malaria patients and healthy individuals as shown in Fig. 3A. The concentration-dependent analysis method showed a correlation coefficient (K2=0.96) between fluorescence intensities and protein concentrations (Fig. 3B).

Optimization of a protein array platform for profiling antibody responses to P. vivax infection. (A) Different concentrations of PvPHIST/CVC-8195-NT/CT and GST protein were probed with pooled vivax patients (P) and healthy individual (H) serum samples. (B) Correlation between spot intensities and the concentration of PvPHIST/CVC-8195-NT/CT and GST proteins. (C) Antibody responses of recombinant PvPHIST/CVC-8195-NT and -CT fragments proteins with vivax patients and healthy individual serum samples by protein array method.
Prevalences of anti-PvPHIST/CVC-8195-NT and -CT antibodies showed that the sensitivity was 61% and 43%, respectively. The sera from vivax patients showed significantly higher MFI than that from healthy individuals. Moreover, NT fragment also performed significantly higher MFI than CT fragment (Fig. 3C, 4). GST alone was not observed to give a significantly high level interaction.
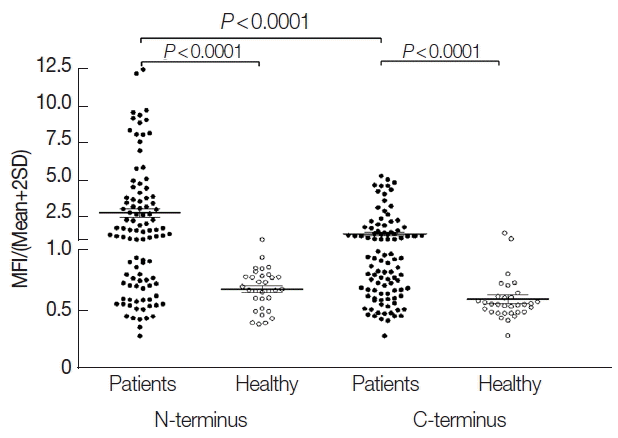
Humoral immune responses to recombinant PvPHIST/CVC-8195-NT and -CT using protein microarrays. Sera from P. vivax-infected patients (n=100) and healthy individuals (n=32) were used for detection of the antibody response to a recombinant PvPHIST/CVC-8195-NT and -CT fragments. Bar indicates the mean+SD. P-value was calculated by the Student’s t-test. MFI, mean fluorescence intensity.
IFA was performed using antisera raised against PvPHIST/CVC-8195-NT and -CT to determine its subcellular location at different stages of P. vivax parasites. The anti-PvPHIST/CVC-8195-NT produced dots spotted across the infected RBC throughout the erythrocytic stages, which is similar to the one shown in P. cynomolgi (Fig. 5A). In contrast to anti-PvPHIST/CVC-8195-NT and -CT fragments, sera presented few or no fluorescence through all erythrocytic stages (Fig. 5B). Anti-GST antibody and preimmune mouse sera were also used as controls. All fluorescence results were negative (data not shown).
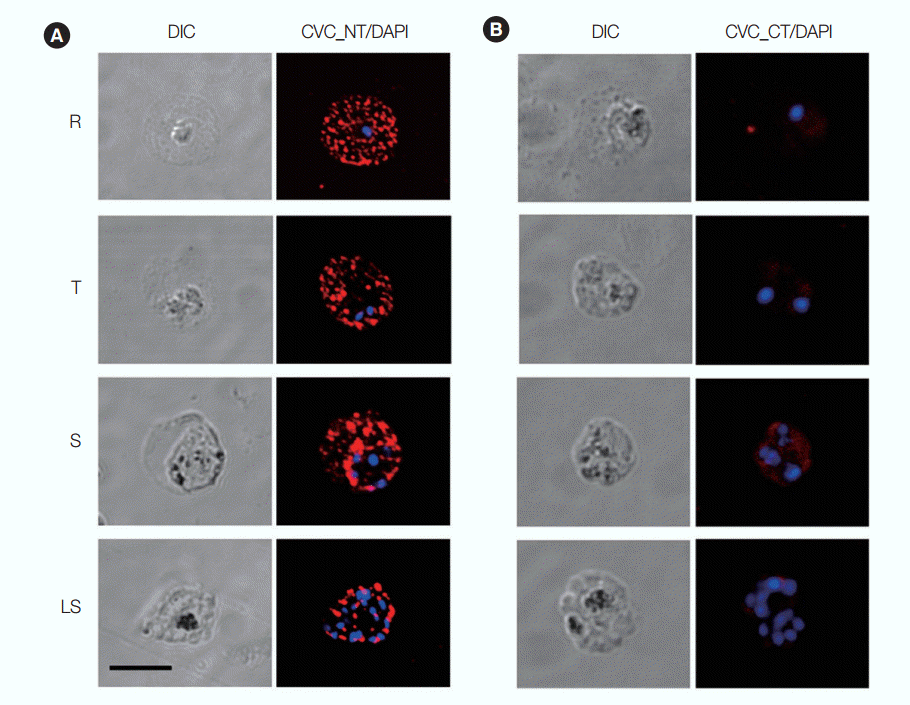
Subcellular localization of PvPHIST/CVC-8195-NT and -CT fragments. Parasites were labeled with antisera against PvPHIST/CVC-8195-NT (A) and -CT (B) at ring stage (R), trophozoite stage (T), schizont and late schizont stages (S and LS). Nuclei are visualized with DAPI in merged images. Bar represents 5 μm.
DISCUSSION
Reticulocytes infected by P. vivax were twisted and increased in size. Three types of unique structures were found in infected reticulocytes compared with uninfected reticulocytes: CVCs, cleft, and the large vacuoles [10]. Among these 3 structures, CVC has never been observed in erythrocytes infected with falciparum-, malariae- or knowlesi-type malaria parasites but existed in erythrocytes infected with P. vivax and P. cynomolgi. The CVCs are composed of alveolar-like clusters of small vesicles that are connected to the base of invaginations or caveolae in the erythrocyte membrane. In this paper, we identified that PvPHIST/CVC-8195 is localized on the CVCs. Furthermore, antigenicity studies confirmed that PvPHIST/CVC-8195-NT were highly antigenic to vivax patients’ serum.
Humoral immune responses to blood-stage malaria antigens are an important component of naturally acquired immunity to malaria [22–24]. Those malaria antigens induced antibodies are likely to play a major role in protection through following mechanisms: prevention of parasite invasion, intra-erythrocytic parasite inhibition, and mononuclear cell-mediated protection [25]. In the current study, IgG antibody prevalence of vivax patients against PvPHIST/CVC-8195-NT showed significantly higher seropositivity and titer than CT fragment.
A recent report suggested that anti-PvPHIST/CVC-8195 antibodies produced a dotted pattern of fluorescence in P. cynomolgi parasites; this dotted pattern was finally proved as CVCs structure in P. cynomolgi parasites. In this study, IFA analysis indicated that anti-PvPHIST/CVC-8195-NT antibodies produced patterns similar to those in P. cynomolgi. So, we inferred that PvPHIST/CVC-8195 was also a CVC associated protein in P. vivax. Meanwhile, anti-PvPHIST/CVC-8195-CT antibodies did not present a CVC associated pattern. Thus, the formation and transport of this protein still need to be further investigated.
In this study, a member of the P. vivax helical interspersed subtelomeric (PHIST) superfamily protein, PvPHIST/CVC-8195 was characterized. The protein expression profile during the P. vivax asexual stage and its CVCs associated localization were determined. The antigenicity of PvPHIST/CVC-8195 was attested by the generation of antibody responses against it during natural P. vivax infection. Further studies on molecular structure and function of this protein might lead to the development of therapeutic strategies against malaria parasites.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2014R1A2A1A11052079), and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (no. 2015R1A4A1038666). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this study.
