Platynosomum fastosum (Trematoda: Dicrocoeliidae) from Cats in Vietnam: Morphological Redescription and Molecular Phylogenetics
Article information
Abstract
The present study was performed to reveal the morphological characteristics and molecular phylogenetic position of Platynosomum fastosum Kossack, 1910. A total 167 specimens of P. fastosum were collected in 8 (4.9%) out of 163 sets of gall-bladders and bile ducts of cats. The number of worms was 1–105 per infected cat. This species was characterized by having a long and slender body, slightly larger ventral sucker than the oral sucker, indistinct prepharynx, small pharynx, short esophagus, bifurcation midway between 2 suckers, and ceca extending to the posterior end of the body. The length of the partial sequences of ITS1 and 5.8S rDNA of P. fastosum were 990 bp, GC-rich. AT/GC ratio was 0.9, there were 9 polymorphic sites, and intraspecific variations ranged from 0.1% to 0.9%. Phylogenetic analyses by neighbor-joining phylogram inferred from ITS1 rDNA sequences revealed that the genetic distance between P. fastosum specimens ranged from 0.3 to 1.5% while the smallest interspecific distance among dicrocoeliid species was 20.9 %. The redescription and genetic characters of P. fastosum are taxonomically important to recognize future different species of the genus Platynosomum showing high intraspecific and morphological variability.
INTRODUCTION
The hepatic trematode, Platynosomum fastosum Kossack, 1910 is one of the most important and widely distributed (geographically) parasites of cats [1]. In Asian countries, this species has been found from domestic cats in Indonesia [2], Korea [3], Malaysia [4,5], Thailand [6,7], and Southern Vietnam [8]. P. fastosum has also been found in wild animals, including the small Asian mongoose, Herpestes javanicus (Saint-Hilaire), in Central Highlands of Vietnam [9]. Additionally, P. fastosum is known to cause pathological symptoms, including cholangiocarcinomas in the liver [10].
P. fastosum was treated to be synonymous with other dicrocoeliid species because of intraspecific morphological variations [11]. Maldonado [12] treated 3 Platynosomum species, i.e., P. fastosum, P. concinnum (Braun, 1901), and P. illiciens (Braun, 1901), recovered in the gallbladder and bile ducts of cats, as the same species. Nguyen and Ha [13] reckoned that Eurytrema rebelle Railliet, 1924 is one of the synonyms of P. fastosum. Some other dicrocoeliid flukes, such as P. planicipitus, Dicrocoelium concinnum, D. lanceolatum var. symmetricum, and Concinnum concinnum, were also considered to be the same species as P. fastosum [1]. Recently, Pinto et al. [11] mentioned that no significant morphological differences were observed between P. fastosum and P. illiciens even though they parasitize different host species; mammals and birds, respectively. There are, however, no molecular studies on Platynosomum spp. up to the present to confirm the opinions of the above authors.
Among all target sequences used for phylogenetic and diagnostic studies, ribosomal DNA (rDNA) is a useful genetic marker for examining taxonomic status of digeneans [14]. Especially, the internal transcribed spacer (ITS1) regions of rDNA may provide useful information for elucidating interspecific and intraspecific relationships among closely related genera in many eukaryotes [15–17].
In the present study, we redescribed to give the species validity of P. fastosum with reinforced morphometric data and provided the phylogenetic data and partial sequences of the ITS1-5.8S rDNA, for more precise characterization of this fluke species. Phylogenetic relationships among the different species of hepatic trematodes (including dicrocoeliid species) were also inferred from the ITS1 rDNA sequences to elucidate their positions.
MATERIALS AND METHODS
Surveyed area and worm recovery
A survey for the recovery of liver flukes was performed with gall-bladder and bile ducts of cats obtained in slaughter houses located in Thai Binh province in Northern Vietnam from April 2012 to July 2015. The gall-bladder and bile ducts of each cat were removed and stored separately in plastic bags, placed on ice, and carried fresh from the slaughter houses to the laboratory. In the laboratory, the samples were necropsied following the method described by Skrjabin [18]. The gall-bladder and bile ducts of the cats were opened by scissors, carefully not to cut too deeply and keep the tip of the scissors pointed upwards and do not damage the underlying structures. After that, these organs were rinsed in the sink and then checked for the presence of liver trematodes under a stereomicroscope Olympus SZ61.
Morphological observations
Trematodes were removed from debris, counted, and identified primarily using the keys by Bray et al. [19] and Nguyen [20]. Some P. fastosum, which were chosen for morphological studies, were transferred to a drop of saline on a microscope slide and covered with a coverslip. The slide and the coverslip were gently rolled by a thread to prevent it from moving. They were fixed in warm AFA (a mixture of 70% ethanol-formalin-acetic acid in the ratio of 90:7:3), preserved in 70% ethanol, stained with Mayer’s carmine, dehydrated using a graded ethanol series (80–100%), cleared in xylene, and mounted in Canada balsam [21]. The morphology of P. fastosum was studied using a light microscope Olympus CH40 at magnifications of ×40 to ×400. Drawings were made with the aid of a camera lucida, and were redrawn using Adobe Illustrator CS6. All measurements are given in micrometers with the range followed by the mean in parentheses. Other P. fastosum specimens were preserved in absolute ethanol for molecular studies.
DNA extraction, PCR, and sequencing
DNA from 3 specimens of P. fastosum, collected from 3 individually infected cats, was extracted using the DNeasy Blood and tissue kit (Qiagen, Hilden, Germany). Partial fragments of ITS1-5.8S rDNA were amplified by PCR using the following primer pair; 5,8SR: 5′-CAT GGC CGC AAT ATG CTT GCA-3′ and 18SF: 5′-CCT GGT AAG TGC AAG TCA GAT GC-3′. PCR was performed in a total volume of 20 μl and contained 0.25 mM of each primer, 1 μl of DNA, 2× Taq PCR Mastermix (Promega, Madison, Wisconsin, USA). Amplification of the partial sequence of ITS1-5.8S was performed on a Eppendorf Mastercycle using the following cycling conditions: 2 min initial denaturation step at 95°C, 35 cycles of 30 sec at 95°C, 30 sec at 53°C and 1 min at 72°C, and a 7-min extension at 72°C. The PCR products of the partial sequences for 3 P. fastosum specimens were sent to First Base Company for sequencing using BD1primer 5′-TCGTAACAAGGTTTCCGTA-3′ [22]. The results of sequencing were checked by eye using Chromas software.
The 3 newly obtained ITS1-5.8S partial sequences of P. fastosum were deposited in GenBank database under accession nos. KU987672-KU987674. These sequences were aligned and examined for variable sites using the MEGA 6.0.6 program [23].
Phylogenetic analyses
Neighbor-joining phylogram inferred from the ITS1 rDNA sequences were constructed with the Kimura-2 parameter model [24] using computer software program package PAUP* 4.0 beta [25]. ITS1 sequences of Fasciola gigantica (acecession no. AB553651) were used as an outgroup for constructing the tree. Additionally, ITS1 sequences of opisthorchiid and dicrocoeliid species were used as references (Table 1). Bootstrap analyses were conducted using 1,000 replicates for a phylogenetic tree. Estimates of evolutionary divergence between the ITS1 rDNA sequences were calculated by MEGA 6.0.6 [23] in consideration with all substitutions and missing and/or gaps as unambiguous changes, and analyses were used the maximum composite likelihood model.
RESULTS
Worm recovery
A total of 163 sets of gall-bladders and bile ducts of cats were examined. P. fastosum was found in 8 cats (4.91%) with 167 specimens, and ranged from 1 to 105 specimens, with a mean of 21 per infected cat. Multiparasitism of hepatic trematodes in cats at study sites was not common, only 2 cats were found infected with 2 species, P. fastosum and Opisthorchis viverrini (Poirier, 1886).
Taxonomic position and morphological descriptions
Family Dicrocoeliidae Odhner, 1911
Genus Platynosomum Looss, 1907
Platynosomum fastosum Kossack, 1910 (Fig. 1)
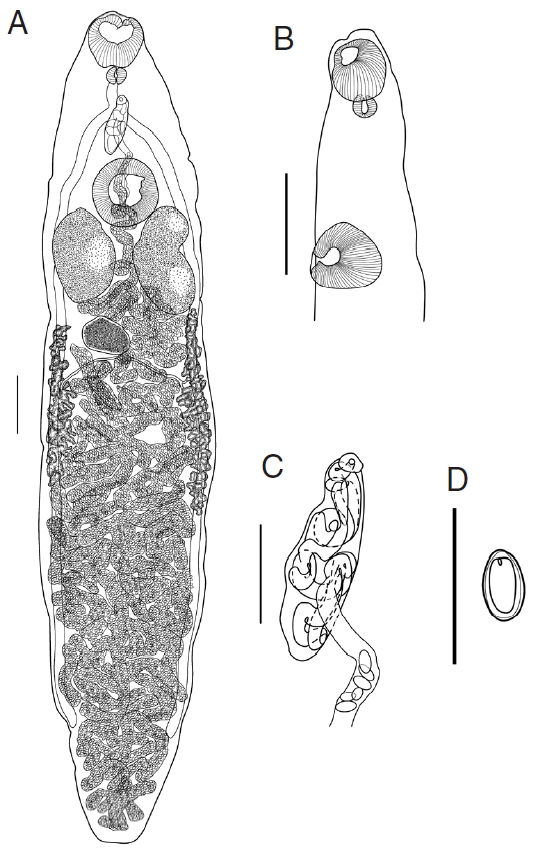
(A) Platynosomum fastosum whole-mount, ventral view. Scale bar=400 μm. (B) Optical sagittal section of the anterior end of body. Scale bar=400 μm. (C) Genital complex. Scale bar=200 μm. (D) Egg. Scale bar=100 μm.
Host: Felis catus
Habitat in host: gallbladder and bile duct of liver
Locality: Dong Hung and Quynh Phu districts (Thai Binh province, Vietnam)
Deposition of specimens: 14 specimens (NMH-Platynosomum-2015-1-14) are deposited in the Department of Parasitology, Institute of Ecology and Biological Resources. Two other specimens are deposited in the Vietnam National Museum of Nature, Vietnam.
Redescription (Based on 16 specimens; measurements in Table 2): Body slender, elongate, with greatest width at mid-length and tapering at both anterior and posterior end; length/width ratio 5.16. Oral sucker muscular, subterminal, more or less spherical. Ventral sucker about 1/4 of the body length from anterior end, transversely oval, protuberant, slightly larger than oral sucker; sucker width ratio 1.09. Prepharynx very short, indistinct, entirely dorsal to oral sucker. Pharynx small, subglobular; oral sucker/pharynx width ratio 2.94. Esophagus short, bifurcates midway between 2 suckers. Intestinal caeca 2, extending towards the posterior end of body, terminate at mid-region of posterior quarter of body.

Morphometrics of Platynosomum fastosum in domestic cats collected in slaughter houses in Northern Vietnam, compared with those reported by previous authors
Testes 2, large, symmetrical, oblong oval, slightly lobed, longer than wide, postero-lateral and positioned on either side of ventral sucker, and separated by coils of uterus. Functional cirrus-sac relatively small, with thin muscular wall, enclosing convoluted seminal vesicle, pars prostatica and short ejaculatory duct, anterior border of ventral sucker. Common genital pore antero-sinistral to intestinal bifurcation.
Ovary entire, sometimes lobed, transversely elongate, smaller than testes, submedian, posterior to 1 of the testes. Seminal receptacle nearly spherical, smaller than ovary, postero-dorsal and positional between 2 testes. The duct of seminal receptacle passes posteriorly and joined by oviduct and then by Laurer’s canal, passes transversely to ovary then joined by duct from vitelline reservoir, then enters Mehlis’ gland beside ovary. Vitellarium follicular; follicles in 2 lateral fields extending from vicinity of posterior margin of testes, surrounding ceca, terminate in mid-region of the 3/4 of body; right band of vitellarium normally shorter than left. Vitelline reservoir immediately posterior to ovary. Uterine coils fill entire hindbody, contain numerous clear to golden brown eggs; anteriorly uterus shows minor coiling in forebody, passes dorsal to ventral sucker, and enters cirrus-sac before opening in the common genital pore.
Sequence and phylogenetic analyses
Partial sequences of ITS1-5.8S of P. fastosum were 990 bp in length, GC-rich; AT/GC ratio was estimated to be 0.9, and nucleotide frequencies were 20.1%, 24.4%, 27.8%, and 27.3% for A, C, G, and T, respectively. A total of 9 polymorphic sites (Table 3) were detected with 5 transitions (A↔G or T→C) and 4 transversions (A→C, A→T, and T→G). The ITS1-5.8S intraspecific variations ranged from 0.1% to 0.9% in the P. fastosum specimens studied.
The neighbor-joining phylogram inferred from ITS1 rDNA comprised 3 lineages of opisthorchiid, dicrocoeliid, and fasciolid species. In the lineage of dicrocoeliid species, all sequences of P. fastosum were formed a monophyletic clade; it was separated with other clades of Dicrocoelium spp. (Fig. 2). The estimation of evolutionary divergence value showed that the genetic distance between P. fastosum specimens ranged from 0.3 to 1.5%, while the smallest interspecific distance among dicrocoeliid species was 20.9%.
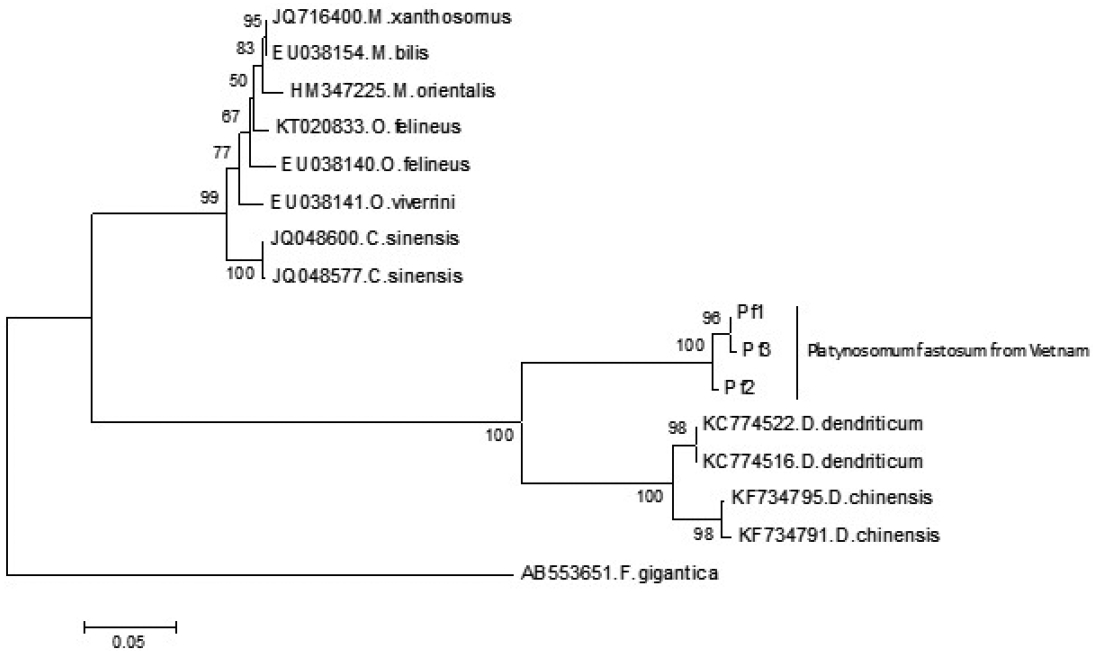
A neighbor-joining phylogram inferred from ITS1 rRNA sequences of Platynosomum fastosum from Vietnam and other related reference flukes in the family Dicrocoeliidae and Opisthorchiidae. The ITS1 rRNA sequence of Fasciola gigantica was used as an outgroup. Bootstrap values higher than 50% are shown on the tree node.
DISCUSSION
Because no local farm produces cat meat in Northern Vietnam, cats are bought from different places and transported to slaughter houses. We have been informed by the slaughter houses that cat sources are mainly from the South of Vietnam, and neighboring countries, such as China, Laos, Cambodia, and Thailand. Cats are purchased through middlemen from various areas and hence their origin is difficult to determine. However, local cats (collected from Thai Binh and several neighboring provinces, e.g., Ninh Binh, Nam Dinh, and Ha Nam) were kept separately in cages, and they were maintained longer because of stronger and little stress after short transportation to compare with other cats.
The coinfection of hepatic trematodes (O. viverrini and P. fastosum) was found only in “long transportation” cats; in 3 infected specimens of local cats only found was P. fastosum. The geographical distribution of O. viverrini is known in Thailand, Laos, Cambodia, and Southern Vietnam [28,29], so the presence of O. viverrini in Northern Vietnam can be explained by transportation of cats from these endemic areas to the slaughter houses/restaurants. The transportation of animals without epidemiological control could be a reason for spread of diseases, including opisthorchiasis from endemic areas to new areas.
Although Pinto et al. [11] and Maldonado [12] believed that P. fastosum is a synonym of P. illiciens, the shape of testes is significantly different between P. fastosum and P. illiciens. The testes of P. fastosum are entire, sometimes slightly lobed, while they are deeply lobed in P. illiciens [27]. This character is sufficiently distinct to justify the distinct species status intermediate within Platynosomum spp. or other taxon. In addition, the body size and length/width ratio of P. fastosum in this study were greater than those in P. illiciens [11]; even other morphometrical measurements were similar between 2 species.
Nguyen et al. [30] found P. fastosum from dogs, but the authors classified it as Eurytrema rebelle. The morphological characters of P. fastosum and E. rebelle are similar; however, the common genital pore of Platynosomum opens at the level of or above the intestinal bifurcation, while it opens posterior to intestinal bifurcation in Eurytrema. Currently, the identification of dicrocoeliid species have been traditionally based on the morphological characteristics of the adult worms, but these criteria of the differentiation are often insufficient for determination; therefore, the molecular genetic approach could be used to identify closely related species.
Up to the present, the applied DNA technology for studying dicrocoeliid species is limited. Only few species have had genetic data, mainly focused on lancet species, i.e., Dicrocoelium spp., and Eurytrema spp. [17,31–33]. In this study, the molecular genetic approach was used to characterize the partial region of rDNA, namely the ITS1 and the 5.8S from 3 P. fastosum specimens. We, however, could not compare the ITS1 sequences with other members of the genus Platynosomum due to unavailability of data in GenBank or elsewhere. The results of our study will be useful for the genetic comparison with other dicrocoeliid species, especially in the genus Platynosomum. Further samples of Platynosomum spp. and other molecular markers are necessary to provide more information for future phylogenetic studies of this genus and genetic diversity in P. fastosum populations within and between geographical regions.
Considering the current taxonomic knowledge on P. fastosum, this species can be distinguished from P. illiciens by only 2 criteria of the class of hosts [11] and the testis shape. The sequencing of molecular markers of P. fastosum and P. illiciens could confirm the non-specificity of P. fastosum or the existence of cryptic species within the genus Platynosomum during their high intraspecific and morphological variability.
ACKNOWLEDGMENTS
We would like to appreciate the valuable contributions of the anonymous reviewers. This research was funded by Vietnam National Foundation for Science and Technology (NAFOSTED) under grant no. 106-NN.05-2014.21.
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.

