Fasciola hepatica: Infection Status of Freshwater Snails Collected from Gangwon-do (Province), Korea
Article information
Abstract
Fasciola hepatica is a trematode that causes zoonosis, mainly in cattle and sheep, and occasionally in humans. Few recent studies have determined the infection status of this fluke in Korea. In August 2015, we collected 402 samples of freshwater snails at Hoenggye-ri (upper stream) and Suha-ri (lower stream) of Song-cheon (stream) in Daegwalnyeong-myeon, Pyeongchang-gun in Gangwon-do (Province) near many large cattle or sheep farms. F. hepatica infection was determined using PCR on the nuclear ribosomal internal transcribed spacer 2 (ITS-2). Among the 402 samples, F. hepatica 1TS-2 marker was detected in 6 freshwater snails; thus, the overall prevalence in freshwater snails was 1.5%. The prevalence varied between collection areas, ranging from 0.0% at Hoenggye-ri to 2.9% at Suha-ri. However, F. gigantica ITS-2 was not detected in the 6 F. hepatica-positive samples by PCR. The nucleotide sequences of the 6 F. hepatica ITS-2 PCR-positive samples were 99.4% identical to the F. hepatica ITS-2 sequences in GenBank, whereas they were 98.4% similar to F. gigantica ITS-2 sequences. These results indicated that the prevalence of F. hepatica in snail intermediate hosts was 1.5% in Gangwon-do, Korea; however the prevalence varied between collection areas. These results may help us to understand F. hepatica infection status in natural environments.
The sheep liver fluke, Fasciola hepatica, is primarily a zoonotic parasite that causes liver infection in cattle and sheep, and occasionally in humans. Freshwater snails are the first intermediate host, and humans are infected mainly by ingesting raw aquatic plants contaminated with the metacercariae [1,2]. F. hepatica infection in snails has traditionally been evaluated by microscopy; however, this method is time-consuming and laborious [2]. In addition, microscopic investigation of snails for F. hepatica larvae has low diagnostic sensitivity [3]. Thus, various molecular approaches have been used to identify Fasciola sp. infectons [3–9].
F. hepatica is distributed mainly in Europe, America, and Oceania [1,2]. The prevalence of F. hepatica infection was high in cattle before 2000 in Korea [10]. There were also several human F. hepatica infection cases [6]. However, little information is available regarding the prevalence of F. hepatica infection in Korea. Therefore, to obtain basic information regarding F. hepatica infection in snail intermediate host in freshwater environments in Korea, we collected freshwater snails from Gangwon-do (Province) and evaluated F. hepatica contamination by PCR.
Freshwater lymnaeid snails were collected at Hoenggye-ri (upper stream) and Suha-ri (lower stream) of Song-cheon (stream) in Daegwalnyeong-myeon, Pyeongchang-gun in Gangwon-do (Province) near large cattle or sheep farms in August 2015 (Fig. 1). A total of 402 samples were collected and examined for the presence of the F. hepatica ITS-2 gene by PCR. Briefly, genomic DNA was extracted from the whole snail body using a G-DEX™ genomic DNA extraction kit (iNtRON Biotechnology, Seoul, Korea) according to the manufacturer’s instructions. Genomic DNA isolated from an adult F. hepatica worm (kindly provided by Prof. Sung-Jong Hong, Chung-Ang University) and adult Fasciola gigantica worm (kindly provided by Prof. Keeseon S. Eom, Chungbuk National University) were used as positive controls. The sequences of the primers used were as follows: F. hepatica ITS-2, forward; GTTATAAACTATCACGACGCCCAAA, reverse; GAAGACAGACCACGAAGGGTA and F. gigantica ITS-2, forward; TATCACGACGCCCAAAAAGT, reverse; CCAAGTTCAGCATCAAACCA. The PCR mixture for amplification contained 5 μl of genomic DNA, 3 μl each of forward and reverse primers, 4 μl of dNTPs, 5 μl of 10× Ex Taq buffer, 0.25 μl of Ex Taq polymerase, and 29.75 μl of double-distilled water (DDW). PCR assays were performed with an initial denaturation step of 94°C for 30 sec, followed by 30 cycles of denaturation at 98°C for 10 sec, annealing at 60°C for 30 sec, and extension at 72°C for 30 sec followed by 1 cycle of 72°C for 10 min and a final hold at 4°C. Amplification was performed using a TaKaRa PCR Thermal Cycler (Takara Bio Inc., Otsu, Japan). Agarose gel electrophoresis (1.5%) with ethidium bromide staining was used to visualize the ITS-2 PCR products.
Map of Daegwalnyeong-myeon, Pyeongchang-gun of Gangwon-do (Province). □Snail collected areas; Hoenggye-ri (upper stream) and Suha-ri (lower stream) of Song-cheon (stream). ♣Large cattle or sheep farms near Song-cheon (stream) of Daegwalnyeong-myeon.
In addition, we confirmed the sequence of PCR products from F. hepatica-positive samples by DNA sequencing analysis. Briefly, after electrophoretic separation, the ITS-2 PCR products were clearly delineated and sequenced directly by SolGent (Daejeon, Korea). The sequences of PCR products were compared with the complete ITS-2 sequences of F. hepatica and F. gigantica obtained from GenBank (accession nos. AJ272053.1 and EU260059.1, respectively), using Clone Manager software (Sci-Ed Software, Cary, North Carolina, USA).
PCR amplification of the positive control (adult F. hepatica DNA) showed a characteristic band at 364 bp for ITS-2. As shown in Fig. 2, the ITS-2 band of F. hepatica was detected in 6 of 402 snails collected from 3 areas, representing an overall F. hepatica infection prevalence of 1.5% (range, 0.0–2.9%); all were dextral. We also performed PCR analysis to evaluate F. gigantica contamination of the 6 F. hepatica ITS-2 PCR-positive samples using F. gigantica ITS-2 gene primers. After PCR, the F. gigantica ITS-2 marker was not detected among the 6 F. hepatica-positive samples. To confirm whether the positive PCR products were from F. hepatica, the complete DNA sequences of the ITS-2 PCR products were compared with that in GenBank (Fig. 3). The results indicated 98.3% similarity among 361 nucleotides between F. hepatica and F. gigantica ITS-2 sequences. The sequences of sample nos. 17, 20, 30, 31, 33, and 36 were 98.1%, 98.1%, 98.6%, 98.3%, 98.1%, 98.6%, and 98.6% similar to the F. gigantica ITS-2 sequence in GenBank (no. EU260059.1), respectively. However, the sequences of sample nos. 17, 20, 30, 31, 33, and 36 showed 99.2%, 99.2%, 99.7%, 99.4%, 99.7%, and 99.2% identity with the F. hepatica ITS-2 sequence in GenBank (no. AJ272053), respectively. Thus, the nucleotide sequences of the ITS-2 PCR products from the 6 F. hepatica PCR-positive samples were 99.4% identical to the F. hepatica ITS-2 sequence in GenBank, but 98.4% similar to the F. gigantica ITS-2 sequences in GenBank. Taken together, the results indicated an overall prevalence of F. hepatica infection in freshwater snails of 1.5%, ranging from 0.0% to 2.9% depending on the collection site (Table 1).
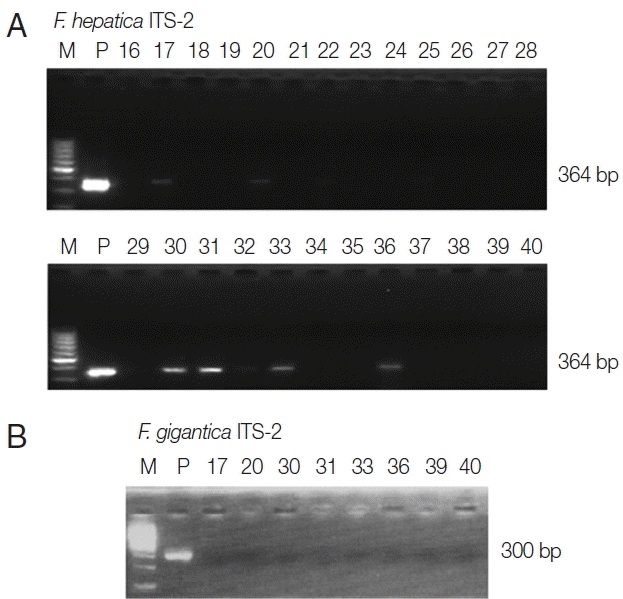
Agarose gel electrophoresis of PCR products, containing the nuclear ribosomal internal transcribed spacer 2 (ITS-2) markers of Fasciola hepatica and Fasciola gigantica. (A) M, 100 bp marker; P, positive control (adult F. hepatica worm); no. 16–40, freshwater snail samples. The total length of the F. hepatica ITS-2 region is 364 bp. (B) M, 100 bp marker; P, positive control (adult F. gigantica worm); no. 17, 20, 30, 31, 33, and 36, F. hepatica ITS-2 PCR positive samples; no. 39–40, F. hepatica ITS-2 PCR positive samples. The total length of the F. gigantica ITS-2 region is 300 bp.

The nucleotide sequences of F. hepatica ITS-2 PCR-positive samples were compared with the F. gigantica ITS-2 (accession no. EU260059.1) and F. hepatica ITS-2 genes (no. AJ272053.1) in GenBank. Base homologies are indicated by a dot (·). Fg ITS2 Eu26, F. gigantica ITS-2 GenBank sequence (no. EU260059.1); no. 17, 20, 30, 31, 33, and 36, F. hepatica ITS-2 PCR-positive samples; Fh ITS2 AJ, F. hepatica ITS-2 GenBank sequence (no. AJ272053.1).
F. hepatica is a snail-transmitted trematode. Approximately 20 species within the family Lymnaeidae play an essential role as the snail intermediate host of F. hepatica worldwide [1,2]. In Korea, lymnaeid snails are distributed throughout the environment; the major habitats are rice paddies, followed by brooks, irrigation canals, and drains [11]. In this study, we used the repetitive DNA sequence of the ITS-2 region specific for F. hepatica to monitor the prevalence of F. hepatica infection. The ITS-2 sequence of nuclear ribosomal DNA was used to identify and determine the genetic variation of F. hepatica [7,9,12–14].
At Daegwalnyeong-myeon of PyeongChang-gun in Gangwon-do, there are many large cattle farms due to well cultivated-grass on large plateau with relatively low temperature. Thus, it would be possible to maintain the life cycle of F. hepatica ecologically. In the present study, we found 6 F. hepatica-infected freshwater snails among 402 snails (1.5%), and all F. hepatica-infected snails were collected at Suha-ri (lower stream), which is the lower part of many cattle or sheep farms, and thus it may be possible to be contaminated with feces of cattle. However, F. hepatica-infected snails were not found at Hoenggye-ri (upper stream), which is located at the upper part of large cattle or sheep farms. The current infection status (1.5%) was lower than the previously reported values of 3.4% in Korea [8], 13.0% in Sweden [12], 7.0% in Switzerland [4], and 26.6% in Poland [5]. In France, the prevalence of natural F. hepatica infection was 1.1% for Lymnaea truncatula and 0.3% for Lymnaea glabra [14].
In addition, to confirm the 6 F. hepatica ITS-2 PCR-positive samples, we performed DNA sequence analysis and compared the results with those in GenBank. The concordance rate of DNA sequences was 99.2–99.7% between the 6 F. hepatica ITS-2 PCR-positive samples and F. hepatica ITS-2 sequences in GenBank. The discrepancy may have been due to point mutation of the ITS-2 gene sequences of F. hepatica in each snail, because the ITS-2 gene was detected in an adult F. hepatica worm as a positive control. In this study, the overall prevalence of F. hepatica infection in freshwater snails was 1.5%, but varied according to locality. These results may help us to understand the F. hepatica infection status of natural environments.
ACKNOWLEDGMENT
The present research has been performed according to the support of Rural Development Administration, Republic of Korea (project no. PJ0108592015).
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
