DNA Methylation of Gene Expression in Acanthamoeba castellanii Encystation
Article information
Abstract
Encystation mediating cyst specific cysteine proteinase (CSCP) of Acanthamoeba castellanii is expressed remarkably during encystation. However, the molecular mechanism involved in the regulation of CSCP gene expression remains unclear. In this study, we focused on epigenetic regulation of gene expression during encystation of Acanthamoeba. To evaluate methylation as a potential mechanism involved in the regulation of CSCP expression, we first investigated the correlation between promoter methylation status of CSCP gene and its expression. A 2,878 bp of promoter sequence of CSCP gene was amplified by PCR. Three CpG islands (island 1–3) were detected in this sequence using bioinformatics tools. Methylation of CpG island in trophozoites and cysts was measured by bisulfite sequence PCR. CSCP promoter methylation of CpG island 1 (1,633 bp) was found in 8.2% of trophozoites and 7.3% of cysts. Methylation of CpG island 2 (625 bp) was observed in 4.2% of trophozoites and 5.8% of cysts. Methylation of CpG island 3 (367 bp) in trophozoites and cysts was both 3.6%. These results suggest that DNA methylation system is present in CSCP gene expression of Acanthamoeba. In addition, the expression of encystation mediating CSCP is correlated with promoter CpG island 1 hypomethylation.
INTRODUCTION
Acanthamoeba cysts are resistant to physical, chemical, radiological conditions, and chemotherapeutic agents [1,2]. Acanthamoeba infection can be difficult to treat due to the cyst stage. In order to understand the encystation mechanism and improve treatment for Acanthamoeba infection, several encystation-mediating factors have been studied. Comparative microarray analysis has shown that 701 genes are up-regulated during the encystation of Acanthamoeba [3]. Autophagosome related proteins, cyst wall related genes, and cyst specific serine and cysteine proteinases have been reported to be needed for the formation of cyst of Acanthamoeba [4–8]. However, regulation mechanisms of these encystation mediating genes are currently unknown.
Gene regulation has a wide range of mechanisms, including increasing or decreasing the production of specific gene products. Gene expression can be modulated by transcriptional initiation, RNA processing, and post-translation of protein. In higher eukaryotes, epigenetic regulation responsible for gene expression has been widely studied. Epigenetics is associated with microRNA expression, DNA-protein interaction, suppression of transposable element mobility, cellular differentiation, embryogenesis, X-chromosome inactivation, and genomic imprinting [9]. Epigenetic modifications can be grouped into 3 main categories: DNA methylation, histone modification, and nucleosome positioning [9]. DNA methylation regulates a number of important biological functions such as chromatin structure, silencing of gene expression, parental imprinting, and chromosome X inactivation [10–12]. Knowledge of DNA methylation and its effects on gene expression in parasites are poorly understood. In protozoan parasite Entamoeba histolytica, 5-methyl cytosine and DNA methyltransferase 2 have been identified [13]. In Plasmodium falciparum, methylated cytosine and a single functional DNA methyltransferase have been reported [14]. Geyer et al. [15] have shown evidence of cytosine methylation in Schistosoma mansoni, suggesting the presence of a functional DNA methylation machinery. However, methylated cytosine and DNA methyltransferase have not been reported in Acanthamoeba yet, although protein arginine methyltransferase 5 has been identified [16].
In this study, we found that 5-azacytidine, an inhibitor of DNA methyltransferase, could reduce the encystation ratio of Acanthamoeba, suggesting that DNA methylation system might be associated with the encystation process in Acanthamoeba. To determine the effect of DNA methylation on gene expression, promoter methylation status and gene expression of cyst specific cysteine proteinase of Acanthamoeba (GenBank accession no. JQ253375) were examined in this study.
MATERIALS AND METHODS
Amoeba cultivation and encystation
Acanthamoeba castellanii Castellani was obtained from the American Type Culture Collection (ATCC 30011). Trophozoites of A. castellanii were axenically cultured in PYG medium (2% proteose peptone, 0.1% yeast extract, and 100 mM glucose) at 25°C in an incubator. Encystation of A. castellanii was induced in an encystation media (0.1 M KCl, 0.008 M MgSO4, 0.0004 M CaCl2, and 0.02 M 2-amino-2-methyl-1,3-propanediol (pH 9.00)) for 3 days.
5-azacytidine demethylation treatment
Acanthamoeba trophozoites were treated with 10 μM of 5-azacytidine (Sigma-Aldrich, St. Louis, Missouri, USA) and incubated in PYG media or encystment media for 3 days. Mature cysts were counted under a light microscope after treating them with 0.5% SDS. Encystation ratios were calculated. Data are expressed as means±SD of the means from 3 independent experiments. Statistical significance was analyzed using unpaired Student’s t-test. A P-value of less than 0.05 was considered statistically significant.
DNA extraction and promoter prediction
Genomic DNA of Acanthamoeba trophozoite was purified using QIAamp genomic DNA kits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Promoter sequences were obtained using GenomeWalker kit (BD Biosciences Clontech, Palo Alto, California, USA), following the protocol recommended by the manufacturer. Briefly, the first step was the construction of pools of adaptor-ligated genomic DNA fragments using 1 of 4 restriction enzymes (EcoRV, DraI, PvuII, or SspI). The first PCR amplification was performed with outer adaptor primer (AP1) provided in the kit and cysteine proteinasegene-specific primer (CP-GSP 1) (Table 1). Nested GSP (CP-GSP 2) (Table 1) and nested adaptor primer (AP2) were used in sequential PCR to amplify the promoter. For primary PCR, the following cycling parameters were used: 7 cycles of 25 sec at 94°C and 4 min at 72°C followed by 32 cycles of 25 sec at 94°C and 4 min at 67°C with a final extension at 67°C for an additional 4 min. Primary PCR products were diluted 1:50 and 1 μl of the diluted primary PCR product was used as template for secondary amplification. Cycling parameters for the secondary PCR were the same as those used for the primary PCR except that the first and second steps consisted of 5 and 22 cycles, respectively. A single band of ~3,000 bp was obtained after PCR amplification. It was cloned into pGEM-T easy vector system (Promega, Madison, Wisconsin, USA). Two clones were picked for plasmid DNA purification for sequencing analysis.
CpG islands and bisulfite sequencing PCR
Promoter CpG islands and bisulfite sequencing PCR primers were designed based on Methprimer site (Table 1). The primers were designed that they didn’t contain any CpGs in order to facilitate their binding to both methylated and unmethylated sequences. Genomic DNA of Acanthamoeba trophozoite and cyst were prepared with QIAamp genomic DNA kits (Qiagen), and Bisulfite kit (Qiagen) was used for bisulfite sequencing PCR. Bisulfite conversion and subsequent purification were performed according to the respective protocols. Seven PCR products (F1–7) were cloned into the pGEM-T easy vector system, and each ten clones were picked for sequencing analysis.
RESULTS
Effect of 5-azacytidine on Acanthamoeba
In order to identify the effect of 5-azacytidine on growth and encystation of Acanthamoeba, trophozoites were cultured with 10 μM 5-azacytidine for 3 days. Results showed that 5-azacytidine had limited effect on the growth of Acanthamoeba compared to untreated control (Fig. 1A, B, E). However, when trophozoites were transferred to encystment media, 5-azacytidine reduced the encystation ratio (Fig. 1C, D, F). Several trophozoites were observed (arrows) after induction into encystment (Fig. 1D). The number of mature cysts was significantly reduced in 5-azacytidine treated cells (52.2%) compared to that in control cells (80.5%) (Fig. 1F). These data indicated that, although 5-azacytidine did not significantly affect Acanthamoeba growth, it reduced encystation ratio.
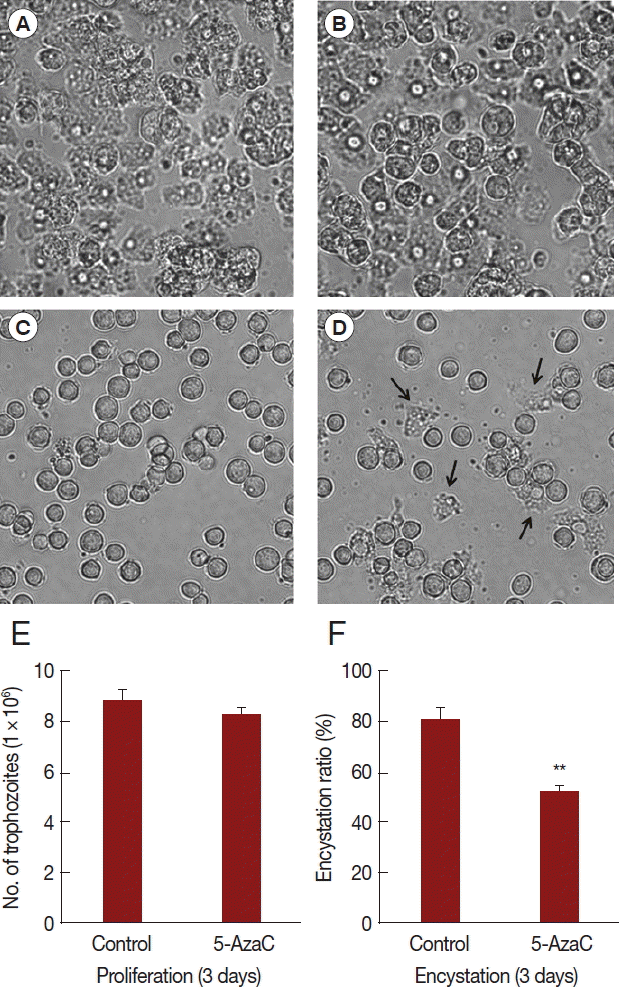
Effect of 5-azacytidine on proliferation and encystation. Acanthamoeba trophozoites were cultured in PYG media without 5-azycytidine (A) or with 5-azacytidine (B) for 3 days. Trophozoites were transferred to encystment media and encystation was induced for 3 days without 5-azycytidine (C) or with 5-azacytidine (D). The 10 μM 5-azacytidine has limited effect on the growth of Acanthamoeba (E). However, encystation ratio was reduced in 5-azacytidine treated cells (52.2%) compared to that in control cells (80.5%) (F). Arrows indicate remained trophozoites after the induction of encystation (D). Values indicate means (±SD) from 3 experiments.
Cloning of cysteine proteinase promoter and prediction of CpG islands
To identify the promoter region of cyst specific cysteine proteinase (CSCP) of Acanthamoeba, a 2,878 bp segment at the 5′-flaking region of CSCP was amplified by PCR using specific primers (Table 1; Fig. 2). CG dinucleotides in the promoter region were underlined. Scanning for CpG islands in the submitted sequence revealed 3 CpG islands in Methprimer (Fig. 3A). The 3 CpG island regions were at least 200 bp with a GC percentage greater than 50%. The observed-to-expected CpG ratio was greater than 60%. The 3 CpG islands were divided into 7 fragments for bisulfite sequencing PCR (Fig. 3B). A total of 14 bisulfite sequencing PCR primers were then designed according to CpG islands (Table 1).

Promoter region of CSCP. Nucleotide sequence of CSCP promoter region was amplified by PCR using GenomeWalker Kit. This DNA is 2,878 bp in length at the 5′-flaking region of CSCP for bisulfite sequencing PCR. CG dinucleotides in promoter region were underlined.
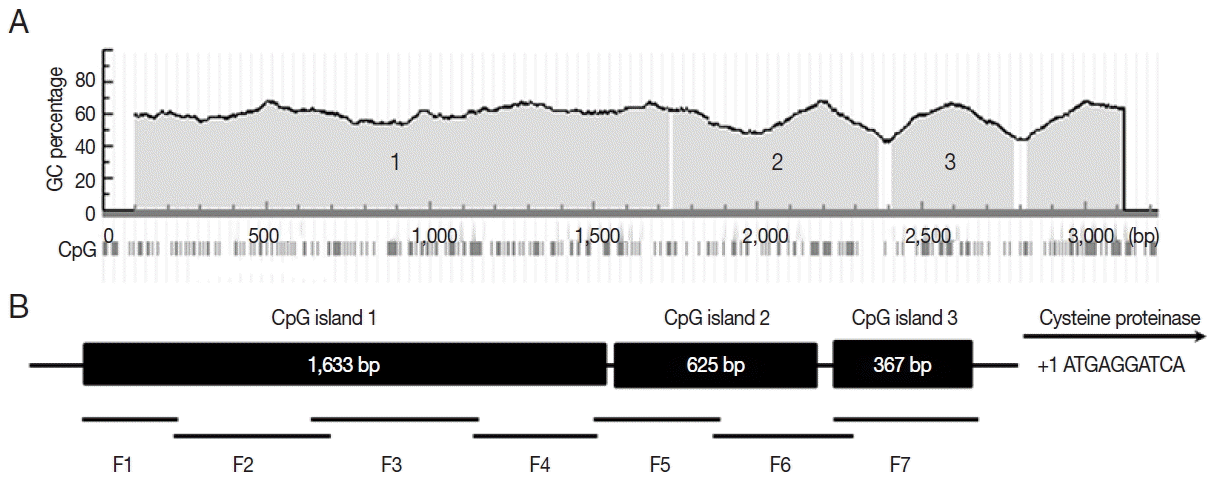
Bioinformatic analysis of CSCP promoter region. Three CpG islands (island 1, 1,633 bp; island 2, 625 bp; island 3, 367 bp) of CSCP promoter region were detected by bioinformatics analysis (A). The promoter region was divided into 7 fragments (B). Primers for bisulfite sequencing PCR were designed based on Methprimer.
Identification of DNA methylation by bisulfite treatment and sequencing
To determine the effect of promoter methylation on gene expression, we performed bisulfite sequencing for encystation mediating CSCP in Acanthamoeba trophozoite and cyst. In trophozoite, 8.2%, 4.2%, and 3.6% methylation were observed in CpG island 1, CpG island 2, and CpG island 3, respectively. At cyst stage, 7.3%, 5.8%, and 3.6% methylation were observed in CpG island 1, CpG island 2, and CpG island 3, respectively. Total promoter methylation status of CSCP in trophozoite was approximately equal to that in cyst. However, CpG island 1 of trophozoite showed higher methylation (8.2%) than that of cyst (7.3%) (Fig. 4). The length of CpG island 1 was 1,633 bp. There were 148 CpG sites.
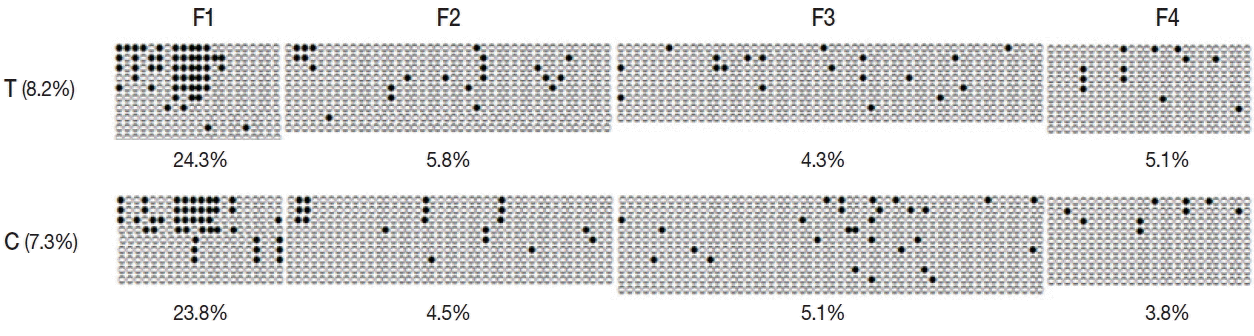
Methylation status of CpG island 1 of CSCP promoter. Methylation of promoter CpG island 1 of CSCP in trophozoite was compared to that in cyst by bisulfite sequencing PCR. In trophozoite CpG island 1, 8.2% methylation was observed while 7.3% methylation was observed in cyst. CpG island 1 of trophozoite showed higher methylation than that of cyst. T, trophozoite; C, cyst; ●, methylated CpG site; and ○, unmethylated CpG site.
DISCUSSION
Cyst specific cysteine proteinase (CSCP) of Acanthamoeba is highly expressed during encystation. The expression of this gene is necessary for encystation of Acanthamoeba [3,8]. In order to understand the regulation of CSCP expression during encystation, epigenetic regulation was examined by analyzing the correlation between promoter methylation status and gene expression in Acanthamoeba.
As an inhibitor of DNA methyltransferase, 5-azacytidine inhibited the formation of mature cyst in Acanthamoeba (Fig. 1). It has been reported that E. histolytica heat shock protein 100 can be induced by 5-azacytidine [17]. In S. mansoni, it has been reported that 5-azacytidine can disrupt egg production and egg maturation [15]. These results suggest that DNA methylation might play essential roles in gene regulation in parasites. Until now, the role of DNA methylation in gene expression of Acanthamoeba has not been reported yet. Therefore, we screened the correlation between DNA methylation and gene regulation in encysting Acanthamoeba.
Here, we showed cytosine methylation in the promoter of CSCP of Acanthamoeba (Fig. 4). CpG island 1 of CSCP promoter in trophozoite showed higher methylation than that in cyst. CpG islands are regions with high frequency of CpG (or CG) sites. CpG island is usually defined as a region of at least 200 bp with GC percentage greater than 50%, and with an observed-to-expected CpG ratio that is greater than 60 %. DNA methylation, occurring in the context of CpG dinucleotide, has profound effect on gene expression by modifying the accessibility of DNA to transcription factors [18].
Although we do not know why CpG island 1 is more effective than other CpG islands, activation of CSCP of Acanthamoeba correlated with CpG island 1 hypomethylation might play a role. Total promoter methylation status of CSCP in trophozoite is approximately equal to that of cyst. Other factors might also affect gene expression of CSCP in encysting Acanthamoeba. However, CpG island 1 was composed of 1,633 bp including 148 CpG sites. CpG island 2 was composed of 625 bp including 45 CpG sites while CpG island 3 was composed of 367 bp including 25 CpG sites (data not shown). Further studies are needed to determine the transcriptional regulation by differential promoter methylation and examine the relationship between promoter methylation status of selected CpG islands in trophozoite and cyst [19].
The detection of methylated cytosine in this study suggests the presence of a DNA methylation system for gene regulation in encysting Acanthamoeba. This also provides an opportunity to investigate the effect of DNA methylation on gene expression associated with encystation. Further studies on epigenetic gene regulation in Acanthamoeba will provide important information to understand gene expression for encystation and amoebiasis control.
ACKNOWLEDGMENT
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2014R1A1A 2058405).
Notes
CONFLICT OF INTEREST
We have no conflict of interest related with this study.
