Simultaneous Molecular Detection of Cryptosporidium and Cyclospora from Raw Vegetables in Korea
Article information
Abstract
Cryptosporidium and Cyclospora are well-known coccidian protozoa that can cause waterborne and foodborne diarrheal illnesses. There have been a few reports regarding contamination in different vegetables with Cryptosporidium, but no data are available regarding the sources of Cyclospora infections in Korea. In the present study, we collected 6 kinds of vegetables (perilla leaves, winter-grown cabbages, chives, sprouts, blueberries, and cherry tomatoes) from July 2014 to June 2015, and investigated contamination by these 2 protozoa using multiplex quantitative real-time PCR. Among 404 vegetables, Cryptosporidium and Cyclospora were detected in 31 (7.7%) and 5 (1.2%) samples, respectively. In addition, Cryptosporidium was isolated from all 6 kinds of vegetables, whereas Cyclospora was detected in 4 kinds of vegetables (except perilla leaves and chives). Cryptosporidium (17.8%) and Cyclospora (2.9%) had the highest detection rates in chives and winter-grown cabbages, respectively. Cryptosporidium was detected all year long; however, Cyclospora was detected only from October to January. In 2 samples (sprout and blueberry), both Cryptosporidium and Cyclospora were detected. Further investigations using TaqI restriction enzyme fragmentation and nested PCR confirmed Cryptosporidium parvum and Cyclospora cayetanensis, respectively. In conclusion, we detected C. cayetanensis in vegetables for the first time in Korea. This suggests that screening should be employed to prevent these protozoal infections in Korea.
INTRODUCTION
Cryptosporidium parvum is a zoonotic protozoan parasite that causes severe enteritis in immunocompromised patients. Numerous outbreaks have been reported worldwide [1–3]. The coccidian parasite Cyclospora cayetanensis has been recently recognized as a cause of prolonged diarrhea in children and immunodeficient patients in developing countries [4–7]. These 2 organisms produce environment-resistant oocysts that are excreted in the feces of infected individuals. The major risk factor associated with transmission of these parasites has been identified as consumption of oocysts in contaminated water or foods [8–11].
Although risks for massive waterborne cryptosporidiosis outbreaks have been present since 1990 in western countries, there was no outbreak in Korea for several decades. However, the first water-borne outbreak of cryptosporidiosis was reported in Korea in 2012 [12], and soil and vegetable samples have been reported to be contaminated with C. parvum oocysts [13]. In contrast, since the first human case of Cyclospora was reported in 2003 in travelers who had visited Indonesia [14], information regarding other indigenous cases and environmental sources of Cyclospora oocysts was not available in Korea. In this study, we investigated several food samples to assess contamination by Cryptosporidium and Cyclospora oocysts using multiplex quantitative real-time PCR (qPCR). This information could be useful for preventing food poisoning due to infections by these protozoa.
MATERIALS AND METHODS
Food sample collection and pretreatment
Six kinds of food samples (perilla leaves, winter-grown cabbages, chives, sprouts, blueberries, and cherry tomatoes) were purchased from local groceries in Seoul from July 2014 to June 2015. For perilla leaves and sprouts, 20 and 30 g samples were used for examination, respectively. For other vegetables, 50 g fresh samples were used. For sample wash preparation, we followed the method reported previously by Hong et al. [13]. Briefly, each sample was placed in a zipper bag (Cleanwrap Inc., Hwaseong, Korea) and 2–3 volumes of 0.1% liquinoxTM (Alconox Inc., New York, USA) in 0.01 M PBS (pH 7.4) was added. Samples were shaken on an orbital shaker (Red Roter, Hoefer, California, USA) for 15 min and were shaken repeatedly in an upside down position. Subsequently, the washing solution was collected and pellets were saved after centrifugation at 5,000 g for 20 min. Food samples were rewashed and pellets were added to the bottle used for the first wash. The pellets were moved into microcentrifuge tubes and kept at −20°C until DNA extraction using QIAquick stool mini kit (QIAGEN Inc., Valencia, California, USA).
All purchased vegetables were produced domestically, except blueberries. Blueberries were imported from the United States (August) and Chile (from December to April) or domestically produced (from May to July).
Real-time PCR
The extracted DNA was used as a template for qPCR. For a positive DNA control, C. parvum oocysts (KKU isolates) were used as described by Lee et al. [15]. The primers and probes used for qPCR were designed using the Swi2/SNF2 ATPase, Rad 16 ortholog gene (Rad 16, GenBank no. XM_625623.1) of Cryptosporidium [13] and the internal transcribed spacer 2 gene (ITS2, GenBank no. AF301386.1) of Cyclospora [16] (Table 1).
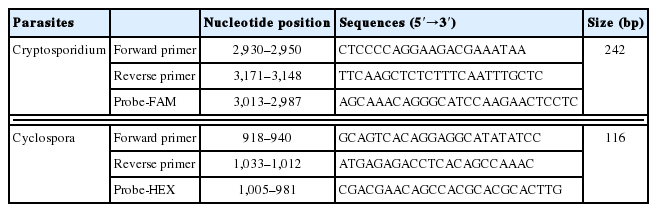
Primers and probes used for multiplex real-time PCR to detect Cryptosporidium and Cyclospora simultaneously in vegetables examined from 2014 to 2015
Absolute qPCR reactions were performed using the method of Lee et al. [15]. Briefly, reaction mixtures included 0.1×LightCycler® FastStart HybProbe master mix (Roche, Mannheim, Germany), each C. parvum primer set at 0.5 μM (Bioneer, Daejeon, Korea), and the probe set at 0.1 μM (TIB MOLBIO, Berlin, Germany). qPCR was performed using a LightCycler® 480 device (Roche), and each qPCR mixture was denatured at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec, and extension at 72°C for 3 sec, with a final cooling step at 40°C for 30 sec. DNase/RNase-free water was used in place of template DNA as a negative control. The plasmid DNA standard for real-time PCR was prepared as described previously using the Rad 16 gene of Cryptosporidium and ITS2 gene of Cyclospora as targets [15].
Sequences of qPCR products were confirmed using an ABI 7700 model sequence detector with SDS v.-1.6.3 software (Applied Biosystems, Foster City, California, USA) at the Cosmo sequence facility service (Seoul, Korea) after electrophoresis using a 2% agarose gel; gene sequences were aligned using Clone Manager suite 7 (Sci-Ed Software, Cary, North Carolina, USA). To differentiate Cryptosporidium species, the qPCR products were digested with the TaqI restriction enzyme (Takara Bio Inc., Shiga, Japan) at 65°C for 2 hr, and analyzed using 2% agarose gels [13]. For Cyclospora species identification, we performed nested PCR as described by Orlandi and Lampel [17] with qPCR positive vegetable samples and the 18S ribosomal DNA gene as the target.
RESULTS
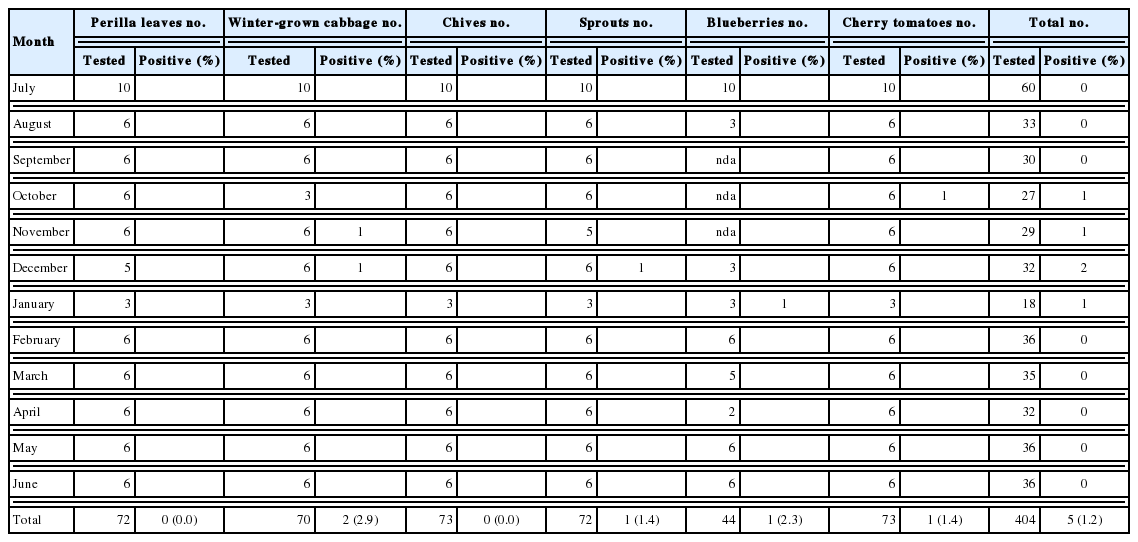
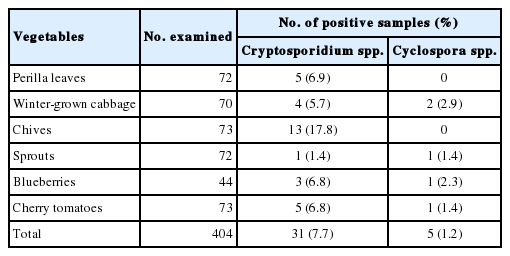
We examined 404 samples representing 6 kinds of popular vegetables to simultaneously detect Cryptosporidium and Cyclospora. As a result, Cryptosporidium spp. was detected in 31 samples of all 6 types of vegetables (7.7%; Table 2). In contrast, Cyclospora spp. were identified in 5 samples comprising of 4 types of vegetables (1.2%; Table 2). The number of oocysts per gram of vegetables ranged from 25 to 783 and from 13 to 348 for Cryptosporidium and Cyclospora, respectively (data not shown).
Positive rates of Cryptosporidium spp. and Cyclospora spp. in various vegetable samples examined from 2014 to 2015
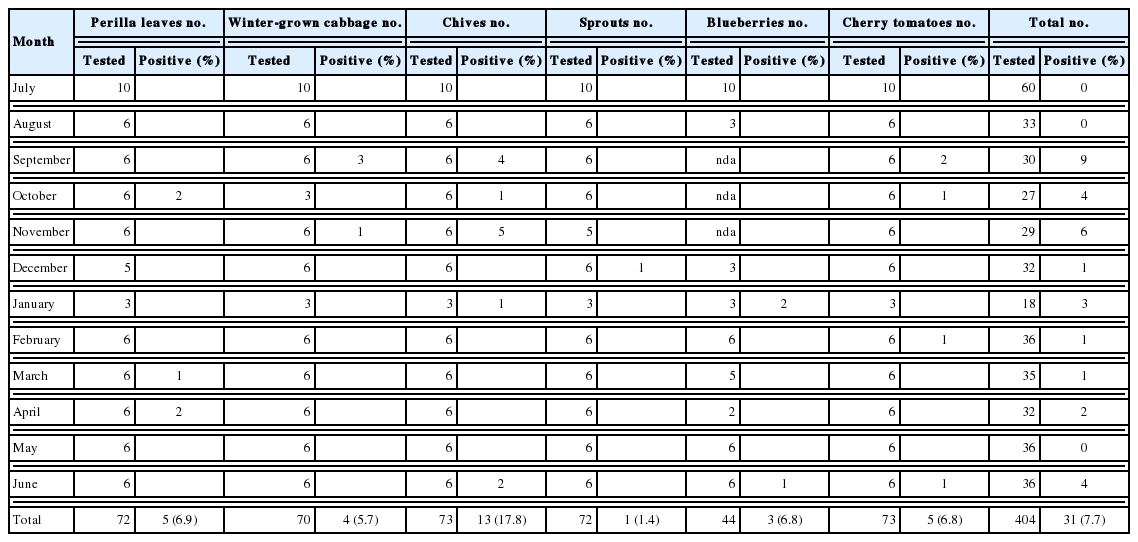
Among the 6 types of vegetables, chives showed the highest contamination rates (17.8%), and sprouts (baby bok choy, chicory, beet, and fushun) showed the lowest contamination rates (1.4%) for Cryptosporidium (Table 3). Perilla leaves, winter-grown cabbages, blueberries, and cherry tomatoes showed a similar range of contamination (5.7–6.9%). Among 3 positive blueberry samples, 2 samples were imported from Chile, and 1 was domestic. In terms of seasonal variation, there was no specific seasonal dependence observed for Cryptosporidium.
Positive rates of Cryptosporidium spp. in various vegetable samples examined from 2014 to 2015 by month
Cyclospora was detected in winter-grown cabbages, sprouts, blueberries, and cherry tomatoes with contamination rates ranging from 1.4% to 2.9% (Table 2). These rates were much lower than those of Cryptosporidium. Especially, both Cryptosporidium and Cyclospora were detected in 1 blueberry and 1 sprout sample. The blueberry sample was imported from Chile, and the sprout sample was a domestic product. In terms of seasonal variation, Cyclospora was detected in samples collected from October to January (Table 4).
TaqI restriction enzyme digestion of qPCR products (242 bp) from Cryptosporidium spp. resulted in 2 bands (117 and 125 bp); therefore, all qPCR positive samples were C. parvum (Fig. 1). We also performed nested PCR targeting 18S ribosomal DNA of Cyclospora using vegetable DNA that was positive for Cyclospora by qPCR as the template. The nested PCR products were 294 bp bands, thus confirming the presence of Cyclospora spp. in these vegetable samples (Fig. 2). Sequencing data from nested PCR products were consistent with the presence of C. cayetanensis.
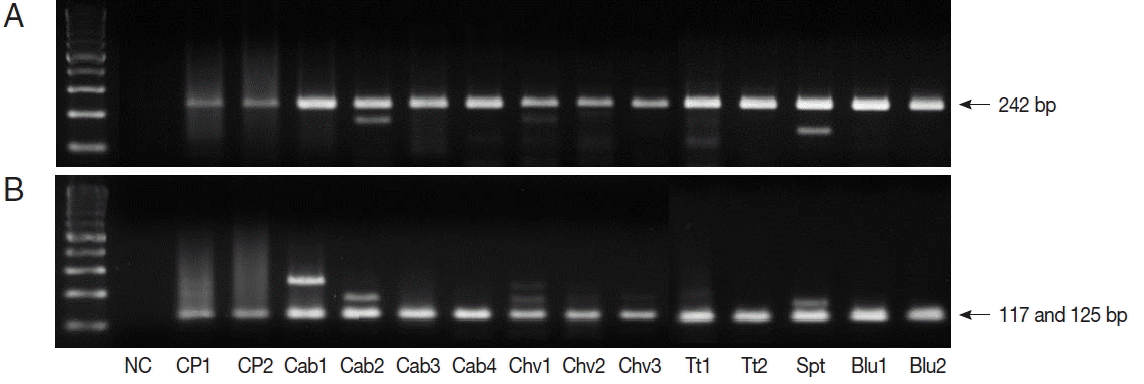
Restriction enzyme fragmentation profiles of Cryptosporidium. Quantitative real-time PCR (qPCR) products in positive samples were cut with TaqI restriction enzyme. (A) DNA of qPCR product. (B) fragmented DNA after TaqI digestion of qPCR product. NC, negative control; CP1 & CP2, C. parvum DNA (for positive controls); Cab, winter-grown cabbages; Chv, chives; Tt, cherry tomatoes; Spt, sprouts; Blu, blueberries.
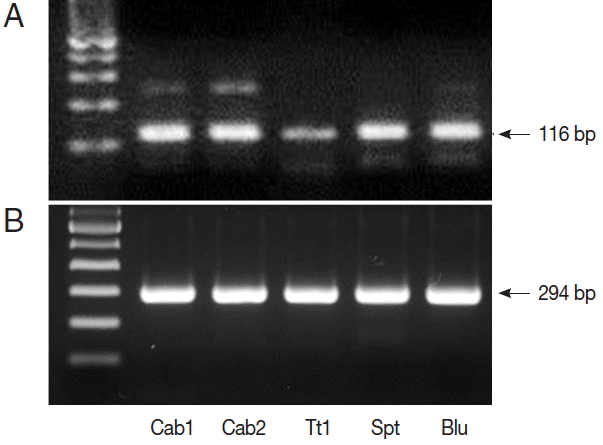
PCR results from Cyclospora positive samples. (A) DNA products after quantitative real-time PCR (qPCR). (B) DNA products after nested PCR. The target gene was internal transcribed spacer 2 (GenBank no. AF301386.1, for qPCR) and 18S rRNA (GenBank no. AF111183.1, for nested PCR). Cab, winter-grown cabbages; Tt, cherry tomatoes; Spt, sprouts; Blu, blueberries.
DISCUSSION
In the present study, we established a qPCR method to detect both Cryptosporidium and Cyclospora simultaneously using fresh farm produce. We identified 2 samples, blueberry and sprout, which were simultaneously contaminated with C. parvum and C. cayetanensis. This technique could be useful to save time and materials, especially those that are available in limited amounts. All vegetable items examined in this study are popular, edible raw, and easily purchased from the grocery markets in town.
The management of opportunistic infections is important because the population of immunocompromised hosts, accompanied by the progress made in transplantation methods, has increased [18]. A recent review article found that in the last decade, only 15 of 71 worldwide Cryptosporidium-linked outbreaks were correlated to foodborne transmission [9]. Reports on foodborne cryptosporidiosis were mainly from the US, UK, and Nordic countries, and various types of food were associated with these outbreaks [19]. In Korea, only 1 waterborne outbreak of Cryptosporidium had been reported until now [12], and there had been no reports of foodborne outbreaks of Cryptosporidium.
In addition, regarding Cyclospora, there had been no cases of indigenous human infections, and there were no reports regarding infection sources in Korea, as only 1 case was reported from an individual infected after visiting Indonesia [14]. It is difficult to explain why cases of cyclosporiasis had not been reported in Korea. This is different from incidence rates in western countries where Cyclospora is considered a major waterborne and foodborne protozoan that is routinely monitored (https://www.cdc.gov/parasites/cyclosporiasis/outbreaks/2016/index.html). Actually, since 1990, more than a half of the foodborne outbreaks by Cyclospora were reported in the US, and several other countries, including Nepal, Canada, Germany, Mexico, Peru, Colombia, Indonesia, and Turkey [20]. Vehicles for most reported outbreaks included various types of berries, salad mixes, and leafy herbs [20]. These are quite similar to the items tested in this study. Perhaps one of the reasons for the absence of cyclosporiasis in Korea is because Cyclospora is frequently neglected as a causative organism of food poisoning. Therefore, it is not screened routinely when foodborne outbreaks occur in Korea.
The increased globalization of the food supply also increases the chances of endemic parasites coming into contact with consumers from other nations. Changes in nutritional habits have resulted in increased consumption of undercooked or raw foods, exposing consumers to parasites that proper food processing would otherwise reduce or eliminate [20,21]. Leafy vegetables are an essential component of a healthy diet but they have been associated with high-profile outbreaks causing severe illnesses [22]. In addition, the consumption of fresh vegetables without proper washing is a risk factor for Cyclospora infection [23].
In the present study, we detected C. parvum and C. cayetanensis in daily edible vegetables, such as winter-grown cabbages, sprouts, blueberries, and cherry tomatoes. This finding is the first report of the infection source of Cyclospora in Korea, whereas C. parvum has been previously reported [13]. Although there have been no reports of Cryptosporidium or Cyclospora foodborne outbreaks, the results suggest that these are possible in Korea. In this report, Cyclospora was detected in samples collected from autumn to winter. The prevalence of Cyclospora infection increased during the warm and rainy season [6,24,25]. In contrast, infection has been reported more prevalent in cool and/or dry season [26,27]. The seasonal trend of Cyclospora infection is not uniform among different regions.
Both Cryptosporidium and Cyclospora can cause infection after consumption of contaminated agricultural water and food [8, 20]. In Korea, Cryptosporidium oocysts were found in 22.5% of water samples from the Han River [28], and C. parvum was reported in 10.5% of pigs in Chungcheongbuk-do [29] and 94% of livestock in Gokseong-gun, Jeollanam-do [30]. Such high livestock infection rates suggest that farming areas nearby stables could be contaminated by agricultural runoff. The finding of vegetables positive for Cyclospora suggests the presence of infected hosts as the source of oocysts, because Cyclospora is an anthroponotic pathogen requiring fecal-oral transmission [20]. A limitation of our study is that although we detected both Cryptosporidium and Cyclospora from several types of fresh produce, it is unknown whether the parasites were infective based on the methods used in this study.
In conclusion, we simultaneously detected C. parvum and C. cayetanensis in several types of fresh produce in Korea using qPCR. Although there have been no foodborne outbreaks caused by these 2 protozoa, this study implies a risk for such occurrences. Therefore, we suggest that fresh vegetables should be carefully washed and care should be taken when eating raw produce to prevent Cryptosporidium and Cyclospora infection, especially in children and immunocompromised individuals. In addition, implementation of monitoring strategies is needed in Korea.
ACKNOWLEDGMENT
This work was supported by Konkuk University in 2014.
Notes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.