PCR Identification and Phylogenetic Analysis of Trichomonas gallinae from Domestic Pigeons in Guangzhou, China
Article information
Abstract
Avian trichomoniasis caused by Trichomonas gallinae is a serious protozoan disease worldwide. The domestic pigeon (Columba livia domestica) is the main host for T. gallinae and plays an important role in the spread of the disease. Based on the internal transcribed spacers of nuclear ribosomal DNA of this parasite, a pair of primers (TgF2/TgR2) was designed and used to develop a PCR assay for the diagnosis of T. gallinae infection in domestic pigeons. This approach allowed the identification of T. gallinae, and no amplicons were produced when using DNA from other common avian pathogens. The minimum amount of DNA detectable by the specific PCR assay developed in this study was 15 pg. Clinical samples from Guangzhou, China, were examined using this PCR assay and a standard microscopy method, and their molecular characteristics were determined by phylogenetic analysis. All of the T. gallinae-positive samples detected by microscopic examination were also detected as positive by the PCR assay. Most of the samples identified as negative by microscopic examination were detected as T. gallinae positive by the PCR assay and were confirmed by sequencing. The positive samples of T. gallinae collected from Guangzhou, China, were identified as T. gallinae genotype B by sequencing and phylogenetic analyses, providing relevant data for studying the ecology and population genetic structures of trichomonads and for the prevention and control of the diseases they cause.
Avian trichomoniasis is a serious protozoan disease mainly caused by Trichomonas gallinae, which infects the upper digestive tract of birds. This parasite infects a wide range of birds, such as pigeons, chickens, turkeys, and other poultry worldwide [1,2]. Infection by T. gallinae may be asymptomatic or may lead to death, with intermediate symptoms, including anorexia, vomiting, ruffled feathers, diarrhea, dysphagia, dyspnea, weight loss, and increased thirst [3–6]. Some reports indicate that multiple large avian mortality events have been associated with trichomoniasis [7,8]. The domestic pigeon (Columba livia domestica) is the main host of T. gallinae and plays an important role in the spread of this disease. Young pigeons are the most frequently infected by this parasite and can die from the infection, but other bird species may act as carriers of the pathogen and show no sign of the disease [9].
In China, pigeon meat is an important part of Chinese cuisine, and nearly 60 million pigeons are raised for human consumption every year [10]. Recent surveys in several provinces of China revealed a high prevalence of T. gallinae infection in domestic pigeons, demonstrating the need to control the spread of the parasite [11–15].
Microscopic morphological examination is the traditional approach used for the identification of T. gallinae, but this approach is labor intensive and requires a specific technique and fresh samples. Previous studies have shown that the internal transcribed spacers (ITS) of nuclear ribosomal DNA (rDNA) provide useful genetic markers that can be used for developing specific PCR assays as an alternative approach for the identification of many parasite species [16–24]. Moreover, sequence analysis has demonstrated that heterogenic species of trichomonads are present in different bird populations and within the same host species. In addition, T. gallinae genotypes A and B and Trichomonas tenax-like strains have been identified in domestic pigeons in Shandong Province, China [15,25], but that information is limited in other areas of China. Hence, the objectives of the present study were to develop a PCR assay for the identification of T. gallinae using genetic markers in the ITS, to compare the results of the PCR assay vs the traditional microscopy method applied to clinical samples collected from pigeons in Guangzhou, South China and to analyze the molecular characterizations of clinical samples.
T. gallinae samples were collected from naturally infected pigeons from a farm in Guangzhou, Guangdong Province, China. Oropharyngeal swab samples of T. gallinae were taken from the mouths of infected pigeons using sterile, pre-moistened, cotton-tipped applicators, confirmed by microscopic examination, and then washed with PBS and centrifuged 3 times, followed by fixation in 80% ethanol for genomic DNA extraction. Other pathogenic organisms infecting pigeons, namely Eimeria spp., Toxoplasma gondii, Cryptosporidium baileyi, Candida albicans, Newcastle disease virus, Escherichia coli, and Salmonella were included as ‘heterologous control’ samples.
Total genomic DNA was extracted from individual T. gallinae samples and ‘heterologous control’ samples by sodium dodecyl sulfate/proteinase K treatment, column-purified using a commercial DNA extraction kit (Wizard DNA Clean-Up System, Promega, Madison, Wisconsin, USA) and eluted into 30 μl of H2O according to the manufacturer’s recommendations [24]. To confirm the identity of the T. gallinae DNA samples, the ITS rDNA sequences were amplified by PCR from each DNA sample using oligonucleotide primers NC5 (forward: 5′-GT AGGTGAACCTGCGGAAGGATCATT-3′) and NC2 (reverse: 5′-TTAGTTTCTTTTCCTCCGCT-3′). Based on the ITS sequences of T. gallinae (see GenBankTM accession nos. JN007005, FN4 33475–FN433477, EU290649–EU290649, EU881911–EU88 1917, and EU215358–EU215369), the following pair of primers was specifically designed to amplify the partial ITS for T. gallinae: TgF2 (5′-AACTAATACCAACTTCTTTT-3′) and TgR2 (5′-TATTCGCGTAGAATAAGAAT-3′). The PCR conditions were optimized for specificity by varying the annealing temperatures and magnesium concentrations. The established PCR amplifications (each in 25 μl) were performed in 10 mM Tris-HCl, pH 8.4, 50 mM KCl, 2.5 mM MgCl2, each dNTP at 250 μM, 50 pmol of each primer, and 0.625 U Taq polymerase (Takara Bio, Osaka, Japan) in a thermocycler (Biometra, Göttingen, Germany).
For PCR, samples were incubated at 94°C for 5 min, followed by 35 cycles at 94°C for 30 sec, 46°C for 30 sec and 72°C for 30 sec, and a final 5 min extension at 72°C to complete the elongation of products. The integrity of the DNA isolated from the ‘heterologous control’ samples was tested using the previously conserved corresponding primers. The specificity of the primers developed for T. gallinae was evaluated using some of the common organisms infecting domestic pigeons, namely Eimeria spp., T. gondii, C. baileyi, C. albicans, Newcastle disease virus, E. coli, and Salmonella, as controls. The smallest amount of DNA detectable by the PCR assay was estimated by a serial titration of genomic DNA from 1 specimen of T. gallinae. A single T. gallinae organism was also placed into a PCR tube and identified by the PCR assay to evaluate its sensitivity. After optimization, the PCR assay was applied to clinical samples collected from a T. gallinae-positive farm in Guangzhou, Guangdong Province, China, and compared with the traditional microscopy method.
The sizes of the PCR products amplified from the DNA samples of T. gallinae and the ‘heterologous control’ samples, using their corresponding primers, varied from 300–1,500 bp on agarose gels in accordance with the expected fragment lengths (not shown), thus demonstrating the integrity of the DNA samples. The optimal annealing temperature was 46°C, and the optimal magnesium concentration was 2.5 mM for the T. gallinae PCR protocol. Under the optimized cycling conditions, the primer sets TgF2/TgR2 amplified a product of approximately 250 bp solely from the T. gallinae samples, whereas no product was amplified from the ‘heterologous control’ samples (Fig. 1). One representative amplicon produced using the PCR assay was sequenced with the corresponding primers and proved to be a partial ITS of T. gallinae (data not shown), thus demonstrating the specificity of the primers, the cycling conditions, and the PCR assay. The smallest amount of DNA detectable by the specific PCR assay was 15 pg. The single organism of T. gallinae could also be detected by this assay (data not shown). These findings showed that the PCR assay developed in this study was both specific and sensitive.
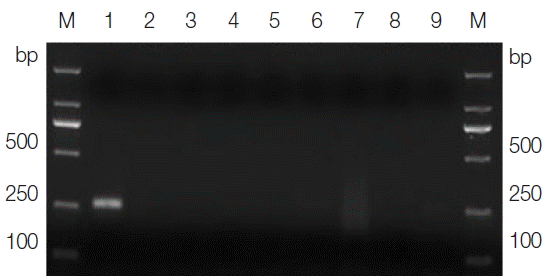
Agarose gel electrophoresis analysis of the specific PCR amplification of ITS for Trichomonas gallinae. Lane 1, genomic DNA from T. gallinae; lanes 2–8, genomic DNA from Candida albicans, Toxoplasma gondii, Cryptosporidium baileyi, Newcastle disease virus, Salmonella, Escherichia coli, and Eimeria sp., respectively; lane 9, no-DNA control. M, DNA size marker (ordinate values in bp).
The PCR assay was used to determine the specific identity of 38 samples collected from pigeons on a T. gallinae-positive farm in Guangzhou, Guangdong Province, China. All of the 15 T. gallinae-positive samples detected by microscopic examination were detected as positive by the PCR assay (data not shown). Moreover, of the 23 samples that were identified as T. gallinae negative by microscopic examination, 19 yielded positive amplifications with the PCR assay. These results were confirmed by sequencing, demonstrating that the PCR assay was more sensitive than the traditional microscopy method. The representative sequences of T. gallinae in this study were deposited in GenBank™ under the accession nos. KY195920–KY19-5926 and were used for constructing a phylogenetic tree. Phylogenetic analyses were conducted using Bayesian inference (BI). Two independent runs were performed for 1,000,000 metropolis-coupled MCMC generations, sampling a tree every 1,000 generations in MrBayes 3.1.1; the first 250 trees represented burn-in, and the remaining trees were used to calculate Bayesian posterior probabilities (Bpp). Phylograms were drawn using the program FigTree v.1.4 (http://tree.bio.ed.ac.uk/software/figtree).
The results showed that T. gallinae from Guangzhou, China, was clustered in the same clade with sequences of T. gallinae genotype B from other geographical origins (Fig. 2). A study on T. gallinae of domestic pigeons revealed the presence of genotype A, genotype B, and T. tenax-like T. gallinae in Shandong Province, China, with genotype A being the most prevalent [15]. There may be differences in the molecular epidemiology of trichomonads in different geographical regions and different bird species. Additional related studies should be conducted elsewhere in China.
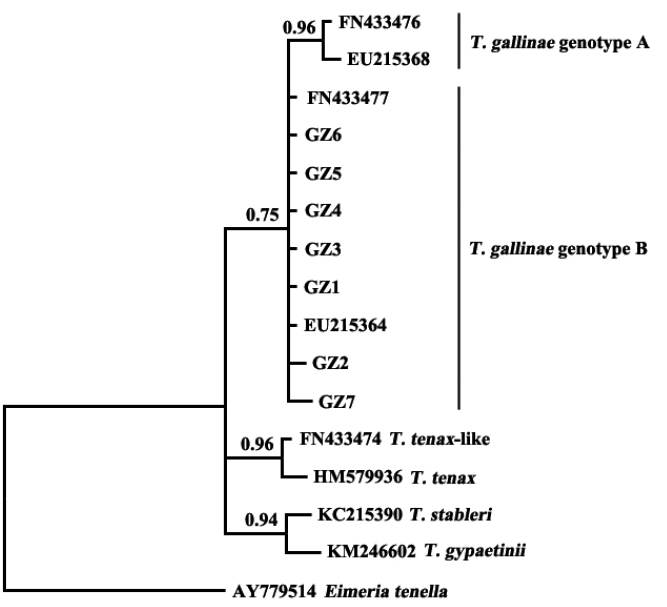
Phylogenetic analyses of trichomonad sequences from the present study and other relevant sequences. The phylogenetic tree was constructed based on partial sequences of ITS1, 5.8S, and ITS2 by the Bayesian inference method.
The present study developed a PCR assay for the diagnosis of T. gallinae infection in domestic pigeons, based on the ITS rDNA sequence as a genetic marker. The developed assay was used to identify the clinical samples and was demonstrated to be a useful tool for the diagnosis and epidemiological investigation of T. gallinae infection. The T. gallinae samples collected from Guangzhou, China, were identified as genotype B by sequencing and phylogenetic analysis, which provided relevant data for further studies on the phylogenetics of trichomonads.
ACKNOWLEDGMENT
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0501303) and the Scientific and Technological Programs of Guangdong Province (grant nos. 2016B020202005 and 2014A020208099).
Notes
CONFLICT OF INTEREST
The authors declare that they have no competing interests.