First Molecular Characterization of Hypoderma actaeon in Cattle and Red Deer (Cervus elaphus) in Portugal
Article information
Abstract
Hypoderma spp. larvae cause subcutaneous myiasis in several animal species. The objective of the present investigation was to identify and characterize morphologically and molecularly the larvae of Hypoderma spp. collected from cattle (Bos taurus taurus) and red deer (Cervus elaphus) in the district of Castelo Branco, Portugal. For this purpose, a total of 8 larvae were collected from cattle (n=2) and red deer (n=6). After morphological identification of Hypoderma spp. larvae, molecular characterization was based on PCR-RFLP and mitochondrial CO1 gene sequence analysis. All larvae were morphologically characterized as the third instar larvae (L3) of H. actaeon. Two restriction enzymes were used for molecular identification of the larvae. TaqI restriction enzyme was not able to cut H. actaeon. However, MboII restriction enzyme differentiated Hypoderma species showing 210 and 450 bp bands in H. actaeon. Furthermore, according to the alignment of the mt-CO1 gene sequences of Hypoderma species and to PCR-RFLP findings, all the identified Hypoderma larvae were confirmed as H. actaeon. This is the first report of identification of Hypoderma spp. (Diptera; Oestridae) from cattle and red deer in Portugal, based on morphological and molecular analyses.
INTRODUCTION
Hypodermosis, one of the most common parasitic diseases in the northern hemisphere [1], is a myiasis caused by warble flies of the genus Hypoderma which is common in Bovidae and Cervidae of the Holarctic region. In Europe, red deer (Cervus elaphus) may be infected by Hypoderma diana and Hypoderma actaeon. H. diana is also found in roe deer (Capreolus capreolus) which is its main host, but H. actaeon seems to be restricted to Cervus elaphus [2].
Some epidemiological studies on hypodermosis have been conducted in wild ungulates in central Europe [3,4], France [5], and southern and central Spain [6,7]. In central Spain, an epidemiological study conducted in red deer was based on the inspection of shot animals, during the period of warble detection. All the third instar (L3) were classified as H. actaeon, and the global prevalence was 61%. The prevalence in yearling and adult deer shot during the official hunting season was 89% [8]. Despite the life cycle of Hypoderma sp. in deer at the western Mediterranean area is not well known, cattle hypodermosis is recognized all over the world [9].
The third instar Hypoderma larvae (L3) can be identified on the basis of their morphological characters. These characters are peritreme structure and the spinulation pattern (presence/absence) in their 10th larval segment [2]. More recently, all 4 Hypodermatinae (H. lineatum, H. bovis, H. diana, and H. actaeon) were accurately characterized by scanning electron microscopy (SEM) by comparing thoracic spines [10]. Additionally, molecular studies on cytochrome c oxidase subunit 1 (CO1) sequences of the mtDNA has been used as a target gene for the identification studies, allowing the distinction of 5 common Hypoderma species together with electron microscopic analysis of key morphological features [11]. Furthermore, Hypoderma spp. have been identified in east Turkey by sequence analysis of mt-CO1 gene and by using PCR-RFLP assay [12].
The identification of Hypoderma larvae described in the literature is scarce, and it is mainly based on the morphology of the 3rd instars [2]. On the other hand, PCR-RFLP and nucleotide sequences of regions of mt-CO1 gene have provided precious information on the classification of Hypoderma spp. [13]. Nowadays, the mt-CO1 gene has been extensively used as a target gene in many identification and molecular phylogenetic studies [14], including Hypoderma spp. [12,15]. This study aimed to identify morphologically and molecularly Hypoderma spp. collected from domestic and wild ruminants in Portugal (cattle and red deer), by morphological characterization, PCR-RFLP analysis, and sequencing analysis of a specific fragment of mt-CO1 gene.
MATERIALS AND METHODS
Study area
The specimens were collected from hunted red deer (6 larvae) and slaughtered cattle (2 larvae) in the region of Castelo Branco (39°8′N; 7°5′W) and Idanha-a-Nova (39°9′N; 7°2′W) during 2011. Idanha-a-Nova and Castelo Branco are 2 municipalities located at the center of Portugal. Every year in this region, hunting activities are organized to control the population of cervids and wild boars. The samples were collected from shot red deer and slaughtered cattle in an effort of adding some more information about this parasite in domestic and wild ruminants. The 2 municipalities are located at 400 m of altitude in a region with a Mediterranean climate with continental influence. The annual average temperature is 15.3°C, and the annual average precipitation 881 mm. July is usual the driest month, and January is the rainiest month.
Sample collection
Hypoderma spp. naturally infected in red deer (Cervus elaphus) and cattle (Bos taurus taurus) were found by the palpation of larvae on the back, flanks, and chest, and by the examination of skin, subcutaneous soft tissues, and subcutaneous muscles, to check for gross changes, such as congestion, bleeding, nodules or lumpy lesions, cysts, and abscesses or suppurative lesions. Larvae were collected, washed several times in PBS (pH 7.4) to remove mucosal debris, preserved in 70% ethanol, and stored at −20°C until analysis.
Morphological identification
The morphological characterization of Hypoderma larvae was performed by using the identification key of Zumpt [2]. Morphological characteristics, such as the shape, size, colour, and pattern of spinulation under a stereomicroscope (Olympus SZX16, Olympus Life Science, Tokyo, Japan) were evaluated. The larval bodies were kept at 70% ethanol until molecular procedures.
Genomic DNA extraction
The total gDNA was obtained from the internal tissue of all individual Hypoderma larvae using commercial Genomic DNA Purification kit® (K0512, Fermentas, Vilnius, Lithuania) with slight modifications. For instance, prior to isolation of gDNA, the ethanol-fixed Hypoderma larvae were washed at least 5 times with saline solution (PBS). The digestion of internal tissues was carried out by the incubation overnight at 56°C with 200 μl lysis buffer provided by the kit to which 20 μl proteinase-K (20 mg/ml) (Sigma, St. Louis, Missouri, USA) was added. After the digestion step, the protocol of the manufacturer was followed. The pellet was resuspended in 100 μl elution buffer, and gDNA were stored at −20°C until further use.
PCR
The amplification of fragments of mt-CO1 gene was done by PCR using conserved specific primers previously described by Otranto et al. [11] (UEA7, 5′-ACAGTTGGAATAGACGTT GATAC-3′ and UEA10, 5′-TCCAATGCACTAATCTGCCATATTA-3′) in a total volume of 50 μl containing 5 μl 10× PCR buffer, 5 μl 25 mM MgCl2, 4 μl of each dNTPs, 20 pmol of each primer, 200 ng of template DNA and 1.25 U of Taq DNA polymerase (MBI, Fermentas). The PCR conditions were 2 min at 95°C (initial denaturation), 40 cycles of 1 min at 95°C, 1 min at 52°C and 1 min at 72°C and finally 7 min at 72°C (final extension). The PCR products were separated on agarose gel (1.4%) and stained with ethidium bromide.
PCR-RFLP
The PCR products were digested at 65°C overnight with restriction endonuclease TaqI, using buffers recommended by the manufacturer (MBI, Fermentas) in 36 μl of the final volume reaction mix, containing 20 μl of PCR product, 10 U enzyme (TaqI, 10 U/μl), 4 μl restriction buffer, and 11 μl distilled water. The restriction fragments were separated on 3% agarose gels and stained with ethidium bromide [11].
For MboII enzyme, PCR products were digested at 37°C during 5 hr, using buffers as recommended by the manufacturer (MBI, Fermentas) in a final 31 μl volume reaction mix, containing 10 μl of PCR product, 10 U enzyme (MboII 10 U/μl), 2 μl restriction buffer, and 18 μl distilled water. The restriction fragments were separated on 3% agarose gels and stained with ethidium bromide. All gDNA positive control samples (previously sequenced) were provided by the Molecular Parasitology Laboratory of Veterinary Faculty, Firat University, Turkey.
mt-CO1 sequence and phylogenetic analysis
All the mt-CO1 PCR products of cattle and deer samples were sequenced. Their nucleotide sequence analysis was undertaken by BLAST algorithms and databases from the National Center for Biotechnology (http://www.ncbi.nlm.nih.gov).
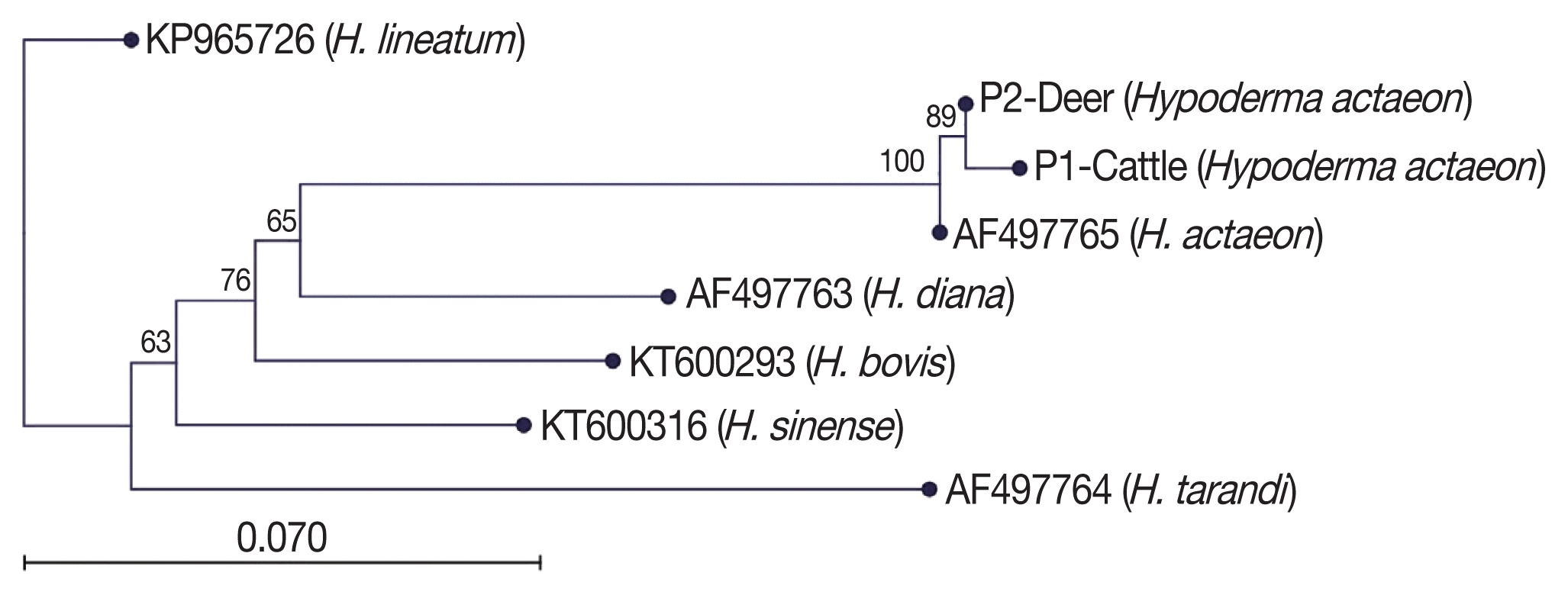
Multiple sequence alignment and phylogenetic tree construction of the obtained sequence of the mt-CO1 gene were performed with the use of CLC Main Workbench software (CLC, Aarhus, Denmark) [16]. Unidirectional DNA sequence analysis of the mt-CO1 gene of all samples were performed. Unreliable ends of the raw sequences were trimmed, and then the phylogenetic tree was built using the neighbour-joining method [17]. Based on pairwise comparisons, sequence differences were calculated using the CLC Main Workbench software. The reliability of the inferred tree was evaluated by bootstrap analysis of 1,000 replicates. Reference sequences of H. bovis (KT600293), H. lineatum (KP965726), H. diana (AF 497763), H. sinense (KT600316), H. tarandi (AF497764), and H. actaeon (AF497765) were also included for phylogenetic analysis.
RESULTS
Morphology
According to the morphological characterization of Hypoderma spp. based on the 3rd instar larvae by stereomicroscope, all of the larvae (n=8) from cattle and red deer were classified as H. actaeon. The spiracular plate of H. actaeon was recessed, and the ecdysal scar well below the surface of the spiracular plate. The evident “C” shaped structures was raised, and the rima was quite open (Fig. 1).
PCR-RFLP
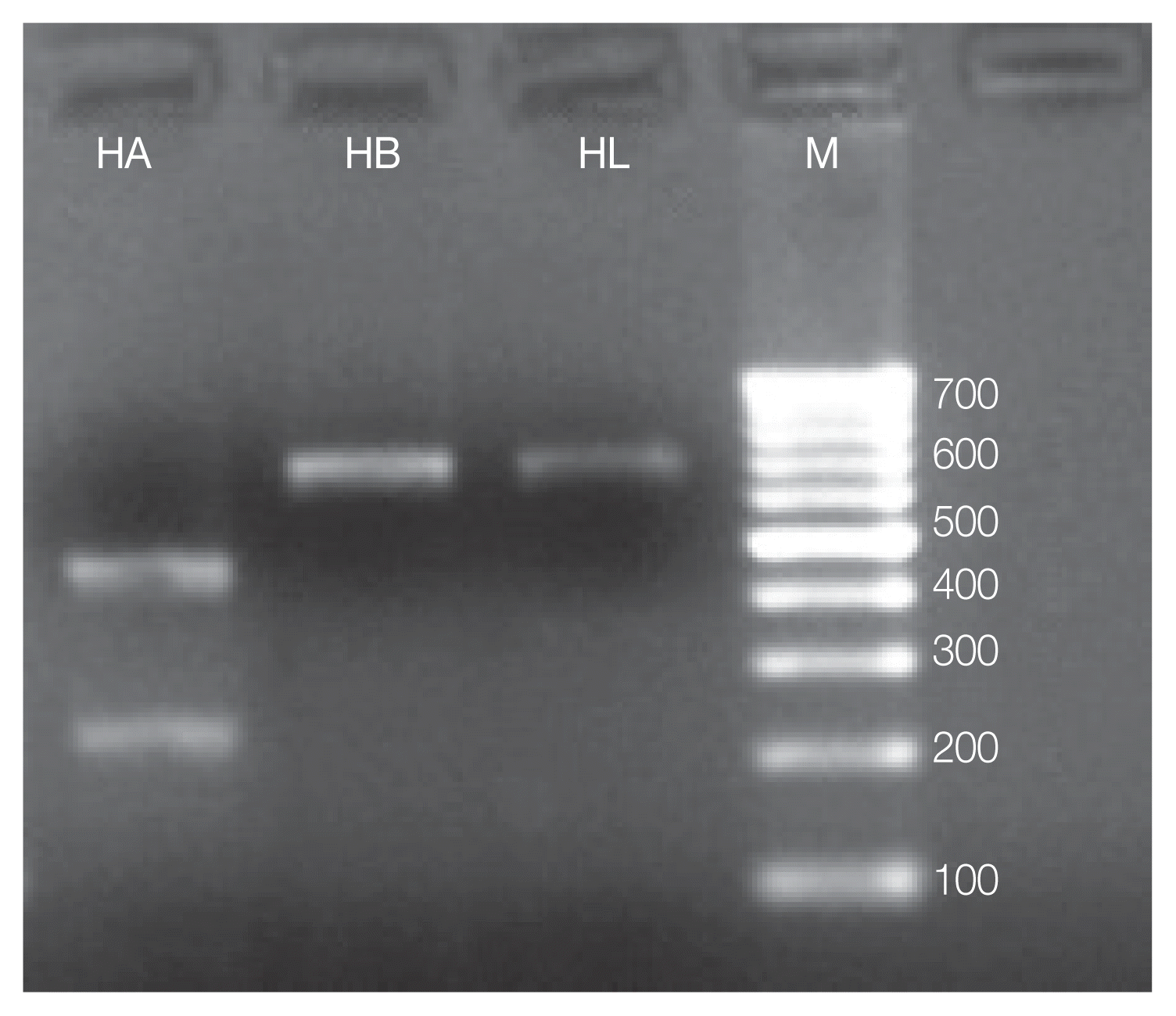
Furthermore, all the specimens were classified as H. actaeon by molecular identification of Hypoderma spp. larvae which was carried out by PCR-RFLP and mt-CO1 gene sequence analysis. Concerning to PCR-RFLP analysis, TaqI restriction enzyme was used to discriminate the Hypoderma species. However, it was impossible to cut fragments of mt-CO1 gene by PCR-RFLP while MboII restriction enzyme was able to discriminate Hypoderma species by the presence of 688 bp bands observed in H. bovis and H. lineatum (no cut), used as a control, and by the presence of 210 and 450 bp bands of H. actaeon (Fig. 2).
mt-CO1 sequence analysis
Blast analysis of Hypoderma spp. sequences in the NCBI was performed and confirmed all the specimens as H. actaeon. We found that these sequences matched with mostly mt-CO1. Then, the sequence results were performed to pairwise analysis for genetic distances. The pairwise comparison and phylogenetic tree obtained from the sequences confirmed that all Hypoderma larvae were classified as H. actaeon (Fig. 3). All sequence results were similar and there were no important variation among the sequences.
DISCUSSION
Hypodermosis shows a worldwide distribution [9] and has been reported in 55 tropical and subtropical countries [18]. This parasitosis is also prevalent in European countries. So far there are limited reports in Portugal despite the high prevalence reported in a neighbour country (i.e. Spain) [19]. Deer hypodermosis is very common in Spain. The prevalence was estimated at 80% [6] and a high intensity of parasite infection was reported (average of 40 larvae) [6,7]. Notwithstanding, more recent studies showed a slight decrease of the prevalence, ranging from 44.8% [20] to 61.0% [8], but the infection intensity remains similar (average of 38.3 larvae) [20].
In wild ungulates, high prevalences of hypodermosis have been reported. In Hungary, 65% of red deer (Cervus elaphus) and 98% of roe deer (Capreolus capreolus) were infected by H. diana while 93% of red deer were infected with H. actaeon [4]. H. tarandi was found in 97% to 100% of Canadian caribou (Rangifer tarandus) [21]. In Europe, H. actaeon is recognized as a typical parasite in red deer [2]; however, an incidence of 89% of hypodermosis in red deer caused by H. diana was reported in Spain [6]. These species are known as strictly host-specific [2]. A seroprevalence study of bovine hypodermosis was conducted in the area of upper Tagus river of Portugal showing a seroprevalence of 35% [22].
This study describes for the first time the presence of H. actaeon in red deer and cattle in Portugal. Regarding the red deer, these findings are in agreement with other studies conducted in Spain where H. actaeon was found in red deer and in roe deer (Capreolus capreolus) from central Spain [7,23]. It was not possible to confirm the presence of H. diana as found in southern Spain [6]. Notwithstanding, the low number of specimens does not allow the exclusion of the possibility of coexistence of these 2 species as it was also observed in Hungary [4].
A limited number of studies based on the morphological identification of Hypoderma larvae from cattle and especially red deer (Cervus elaphus) are available in the literature. These were mainly based on the morphological features of the 3rd instars [2]. In the present study, all the Hypoderma larvae were morphologically identified as H. actaeon. Regarding the deer hypodermosis, H. diana and H. actaeon are the main species that infect deer. These 2 species are morphologically different, since H. diana has a distinctive band of spines on the cephalic segment, which are absent in H. actaeon. These differences facilitate the identification of these species [10].
Nevertheless, morphological discrimination of Hypoderma spp. is not considered an absolutely reliable methodology for identification. Hypoderma from different geographical origins may present only slight morphological differences. Similarly, it is very difficult to differentiate H. lineatum, H. bovis, and H. actaeon from cattle and red deer due to the colour and spinulation patterns. Other difficulties in morphological identification might be due to inappropriate preservation and dark brown colour of the larvae. A more reliable method to identify Hypoderma larvae is using PCR-RFLP based on mt-CO1 gene and sequence analysis [24]. The molecular identification based on mt-CO1 gene has been used to differentiate the 5 common Hypoderma spp. (i.e., H. diana, H. bovis, H. lineatum, H. actaeon, and H. tarandi) [11].
In the present study, a comparative differentiation of H. bovis and H. acteon was done by using PCR-RFLP based on TaqI and MboII restriction enzymes. It reflects the significance of PCR-RFLP method for the accurate identification of Hypoderma spp. in cattle and red deer. The molecular characterization of Hypoderma spp. (H. bovis, H. lineatum, and H. sinense) was based on mt-CO1 gene [25]. According to the mt-CO1 gene sequences analysis and PCR-RFLP findings, all the larvae were molecularly characterized as H. actaeon confirming the results of the morphological identification. Our findings on molecular characterization are similar to those reported by Otranto et al. [11,26]. In Portugal, no previous studies based on morphological and molecular characterization of Hypoderma spp. are available.
A partial mt-CO1 gene was successfully used for amplification of the Hypoderma spp. The gene segment was sequenced and compared to previously determined sequences of H. bovis and H. lineatum. The evolutionary rate of different insects (order; Orthoptera) has been determined by using mt-CO1 gene [27]. The evolutionary rate of different insects (Orthoptera, Diptera, and Hymenoptera) has been determined by using mt-CO1 gene [14,27]. To define the relationships of H. sinense to H. lineatum and H. bovis, the mt-CO1 gene was used as a target for sequence analysis. The clear differentiation of Hypoderma species (H. sinense from H. lineatum and H. bovis) was done by comparing the mt-CO1 gene sequences.
In conclusion, our findings allowed us to state confidently that H. actaeon is an occurring species in red deer and cattle in Portugal. PCR-RFLP is recommended for species identification as compared to sequencing. This study opens new directions of research on Hypoderma spp. in different hosts and different geographical areas of Portugal.
ACKNOWLEDGMENT
We are very thankful to TUBITAK (2216-research fellowship program for international researchers) for providing an opportunity and funding to the work developed by Dr. Haroon Ahmed.
Notes
CONFLICT OF INTEREST
The authors fully declare no financial or other potential conflict of interest.