Molecular Prevalence of Cryptosporidium spp. among Companion Birds Kept in Pet Shops in Japan
Article information
Abstract
Cryptosporidium is the most common protozoan that can infect a wide range of animals, including mammals and birds. Avian Cryptosporidium spp. can cause enteric and respiratory diseases which can be fatal in birds and some species are zoonotic. Companion birds have the potential as reservoir due to their close contact with humans. Pet shops are the major source of companion birds. However, few reports are available regarding Cryptosporidium spp. infection among companion birds kept in pet shops. The present study reports the prevalence and molecular characteristics of Cryptosporidium spp. among companion birds kept in pet shops in Japan. A total of 265 fresh fecal samples were obtained from birds kept in 4 pet shops; these birds belonged to 41 species in 3 bird orders. A nested polymerase chain reaction (PCR) assay targeting the small subunit rRNA gene was employed for the detection of Cryptosporidium spp. A total of 24 samples (9.1%) were positive, and Cryptosporidium spp. were detected from all pet shops. The prevalence of Cryptosporidium spp. in each of the bird orders was 6.5% (10/153) in Psittaciformes, 14.4% (13/90) in Passeriformes, and 4.5% (1/22) in Galliformes. Based on sequence analysis, 13 (54.2%) isolates were classified to C. galli, 8 (33.3%) were avian genotype III, and the remaining 3 (12.5%) were C. baileyi. No infection with zoonotic C. meleagridis and no coinfection with multiple Cryptosporidium spp. and/or genotypes were observed. The zoonotic potential of Cryptosporidium spp. infecting companion birds kept in pet shops in Japan is likely to be low.
The parasite Cryptosporidium is one of the most common protozoans that can infect a wide range of animals, including mammals and birds worldwide [1]. In birds, cryptosporidiosis, which is often fatal, is mainly induced by the 3-dominant species: C. meleagridis, C. galli and C. baileyi. C. meleagridis and C. galli infect the gastrointestinal tract and cause enteritis [2,3], whereas C. baileyi can infect many organs and mainly causes respiratory disorders [3]. In addition, Cryptosporidium avian genotypes III and V have the pathogenic potential to cause mortality, weight loss, chronic vomiting and diarrhea in birds [4–6]. Many other species and genotypes were also identified in birds [3,5]. Among the avian isolates of Cryptosporidium spp., C. meleagridis can infect humans and cause digestive tract obstruction in immunocompromised adults and children [7–10]. In some locations, approximately 10% of human cryptosporidiosis is caused by C. meleagridis [8]. Therefore, Cryptosporidium spp. infection in birds has pathogenic significance for birds and zoonotic risk for humans. In particular, companion birds have a considerable potential to serve as a reservoir due to their close contact with humans. However, the knowledge about Cryptosporidium spp. infection in companion birds is limited, and no report exists regarding Cryptosporidium spp. infection among companion birds kept in pet shops in Japan, although pet shops are the major source of companion birds for private owners. The present study reports the recent prevalence and molecular characteristics of Cryptosporidium spp. among companion birds kept in pet shops in Japan.
Between March 2015 and May 2016, a total of 265 fresh voided fecal samples were collected on a single occasion from each birdcage (birds of a single species were kept in each cage) in 4 pet shops (Shop 1–4) located in the Kanto region, which includes the prefectures of Tokyo (Shop 1; n=78), Chiba (Shop 2; n=53), Saitama (Shop 3; n=59), and Gunma (Shop 4; n=75), in Japan. All pet shop managers granted permission to include their birds in the examination. The birds belonged to 41 species in 3 orders (Psittaciformes, Passeriformes, and Galliformes). The fecal samples were collected immediately after natural defecation and were stored at 4°C prior to DNA extraction (within 3 days). The extraction of Cryptosporidium spp. DNA was performed using a QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions, and the extracted DNA samples were stored at −20°C prior to analysis.
A nested polymerase chain reaction (PCR) assay targeting the small subunit (SSU) rRNA gene was employed for the detection of Cryptosporidium spp. In the primary reaction, the forward primer (5′-TTCTAGAGCTAATACATGCG-3′) and the reverse primer (5′-CCCATTTCCTTCGAAACAGGA-3′) were used to amplify a DNA fragment of approximately 1,325 bp, and in the secondary reaction, the forward primer (5′-GGAAGGGTTGTATTTATTAGATAAAG-3′) and the reverse primer (5′-AAGGAGTAAGGAACAACCTCCA-3′) were used to amplify a fragment of approximately 826 bp [11]. For the primary reaction, the PCR mixture comprised a 1×buffer containing 1.5 mM MgCl2, 200 μM aliquots of each dNTP, 0.5 μM aliquots of each primer, 1.25 units of GoTaq DNA polymerase (Promega Corporation, Madison, Wisconsin, USA), and 3.0 μl of template DNA in a total reaction volume of 25 μl. For the secondary reaction, the PCR mixture was the same as that for the primary reaction, except the amplicons from the primary PCR reaction were used as the template. The following cycling parameters were used for the primary reaction: after an initial denaturation of 3 min at 95°C, 35 cycles were performed, each consisting of 45 sec at 95°C for denaturation, 45 sec at 59°C for annealing, and 60 sec at 72°C for extension, followed by a final extension step of 5 min at 72°C. The cycling parameters for the secondary reaction were as follows: an initial denaturation step of 3 min at 95°C; 35 cycles of 30 sec at 95°C, 60 sec at 58°C, and 1 min at 72°C; and a final extension step of 5 min at 72°C.
All secondary PCR products were identified by electrophoresis on 1.5% agarose gels. The specific DNA fragments (approximately 826 bp in length) were confirmed by alternative ethidium bromide staining and visualization under UV light using a transilluminator. Secondary PCR amplicons of the predicted size were purified using a QIAquick Gel Extraction kit (QIAGEN GmbH, Hilden, Germany) and sequenced with the primer set used in the secondary PCR reaction. Sequences were analyzed by a commercial laboratory (Takara Bio Inc., Kusatsu, Shiga, Japan). Sequence alignment and compilation were performed using the MEGA 6.06 (www.megasoftware.net) program. To determine the species and/or genotypes of Cryptosporidium, the DNA sequences were compared to GenBank references by BLAST searches (http://www.ncbi.nlm.nih.gov/), and the similarity between the isolated and reference sequences was determined based on the degree of sequence identity.
Among the 265 fecal samples from the companion birds kept in pet shops, 9.1% (24 samples) were positive for Cryptosporidium spp., as determined by conventional PCR (Fig. 1). Cryptosporidium spp. were isolated from birds in each pet shop, with a prevalence of 5.3–13.2% (Table 1). Of the 41 examined bird species in the 3 orders, 10 of the bird species belonging to one of the 3 orders were positive for Cryptosporidium spp. (Table 2). The prevalence of Cryptosporidium spp. in each of the 3 orders of birds was 6.5% (10/153) in Psittaciformes, 14.4% (13/90) in Passeriformes, and 4.5% (1/22) in Galliformes. In the order Psittaciformes, birds of 5 bird species were positive for Cryptosporidium spp.: the Rosy-faced lovebird (17.2%, 5/29), the Lilian’s lovebird (20.0%, 1/5), the barred parakeet (100%, 2/2), the Pacific parrotlet (33.3%, 1/3), and the cockatiel (10.0%, 1/10). In the order Passeriformes, birds of 4 species were determined to be infected with Cryptosporidium spp.: the society finch (33.3%, 5/15), the Java sparrow (4.0%, 1/25), the zebra finch (20.0%, 4/20), and the domestic canary (13.6%, 3/22). The only species in the order Galliformes that tested positive was the chicken (6.7%, 1/15).
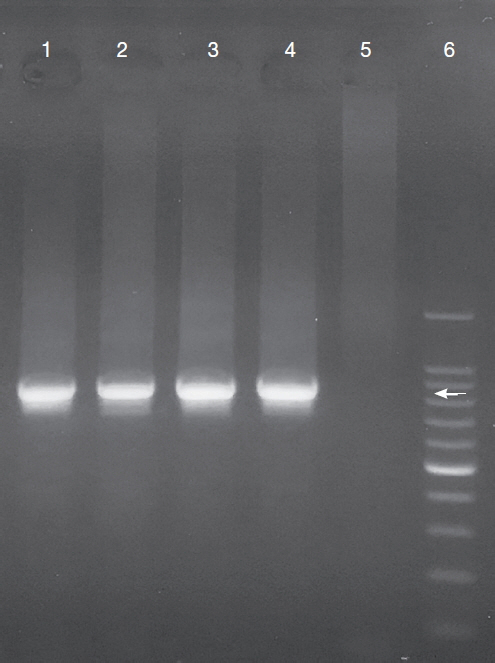
PCR products on 1.5% agarose gel. Lane 1: Cryptosporidium positive control (C. canis), lane 2: C. baileyi from domestic canary, lane 3: C. galli from barred parakeet, lane 4: Cryptosporidium avian genotype III from Lilian’s lovebird, lane 5: Cryptosporidium negative control, lane 6: 100 bp DNA ladder. ←indicates approximately 826 bp.
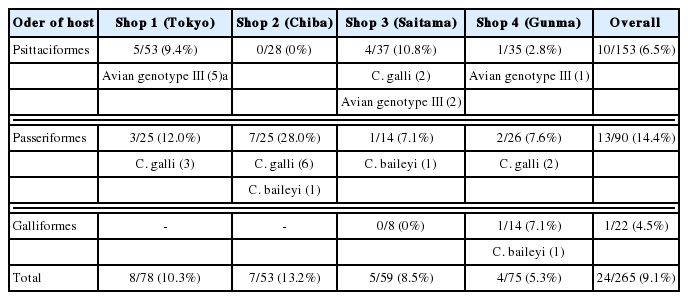
Molecular prevalence and characterization of Cryptosporidium spp. among companion birds kept in pet shops
Based on sequence analysis, 2 species and one genotype of Cryptosporidium were identified. The identity of the 24 positive samples was determined as follows (sequence similarity 99.2–100%): 13 (54.2%) isolates were C. galli (accession number KY490554), 8 (33.3%) isolates were avian genotype III (accession number AB694729), and the remaining 3 (12.5%) isolates were C. baileyi (accession number KU744846) (Fig. 2). No infection with C. meleagridis and no coinfection with multiple Cryptosporidium spp. and/or genotypes were observed. C. galli was found in birds from all pet shops (Shops 1, 2, 3, and 4). Avian genotype III was detected in 3 pet shops (Shops 1, 3, and 4), and C. baileyi was also detected in 3 pet shops (Shops 2, 3, and 4). Thirteen C. galli isolates were found in 5 bird species. One of these species was the barred parakeet (2 isolates), in the order Psittaciformes. The other 4 bird species were the society finch (5 isolates), the Java sparrow (1 isolate), the zebra finch (4 isolates), and the domestic canary (1 isolate), all of which belong to the order Passeriformes. Eight positive isolates of avian genotype III were found in 4 bird species; the Rosyfaced lovebird (5 isolates), the Lilian’s lovebird (1 isolate), the Pacific parrotlet (1 isolate), and the cockatiel (1 isolate), all of which belong to the order Psittaciformes. Three C. baileyi isolates were identified in 2 bird species. One of these species was the domestic canary (2 isolates), in the order Passeriformes; the other was the chicken (1 isolate), in the order Galliformes.
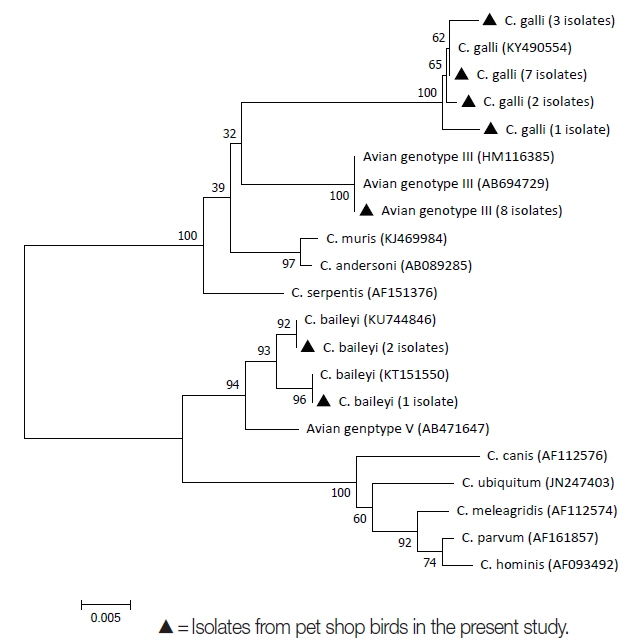
Phylogenetic analysis of the small subunit rRNA gene sequences from Cryptosporidium spp. isolates among companion birds kept in pet shops in Japan.
The present study is the first report to demonstrate the prevalence of Cryptosporidium spp. among companion birds kept in pet shops in Japan. Information regarding the prevalence of infection by Cryptosporidium spp. among birds kept in pet shops and/or bird kept in markets is limited [12–16]. Previously, the prevalence has been reported as 6.8% (7/103) in birds kept in bird markets and pet shops in Brazil [13] and 3.2–13.4% from birds kept in bird markets or pet shops in China [12,14–16]. Due to differences in research methodology, population and scale, comparison between the present study and previous reports is difficult. However, the present results suggest that infection with Cryptosporidium spp. is infrequent but widespread among companion birds kept in pet shops in Japan because isolates of Cryptosporidium spp. were obtained from companion birds in all pet shops and bird orders examined. In the present study, 10 bird species belonging to 3 orders were found to be infected with Cryptosporidium spp. Since these species are common as companion birds, Cryptosporidium spp. infections have previously been detected in the same or closely related bird species [12–20].
In the present study, C. galli was found in the orders Psittaciformes and Passeriformes, and most of the isolates were from the order Passeriformes. Avian genotype III was limited to the order Psittaciformes. C. baileyi was found in the orders Passeriformes and Galliformes. In contrast, 2 or 3 species of Cryptosporidium were confirmed in all shops. Previous reports demonstrated that C. galli has been isolated from the bird orders Psittaciformes, Passeriformes, Galliformes, and others [5,14,15]. Avian genotype III was isolated from the bird orders Psittaciformes and Passeriformes [5,14]. C. baileyi was infrequently isolated in the present study, but was determined to infect quite a wide range bird orders, including Psittaciformes, Passeriformes, and Galliformes [3,5,14,15]. Avian genotypes III, V, and C. galli have been detected in companion birds of the order Psittaciformes kept in private households in Japan [4,6,17]. Additionally, C. baileyi has been isolated from Passeriformes [6]. Therefore, the present study is the first to identify C. galli in Passeriformes and C. baileyi in Galliformes among pet birds in Japan. Although C. meleagridis was not detected here, this zoonotic species was isolated from companiont birds kept in private households in Japan [6,17]. Thus, potential for zoonotic transmission of C. meleagridis from companion birds kept in pet shops to humans is not negligible.
Although the pathogenic potential of the species and genotypes of Cryptosporidium isolates in the present study was indicated [2–6], the infected birds exhibited no clinical signs, with the exception of one Pacific parrotlet (infected with avian genotype III) that displayed anorexia. These results suggested that many companion birds kept in pet shops infected with Cryptosporidium spp. display subclinical infection, and those carrier birds have a risk of developing to clinical cryptosporidiosis in future.
Notes
CONFLICT OF INTEREST
On behalf of all authors, the corresponding author states that there is no conflict of interest.