Survey and Phylogenetic Analysis of Rodents and Important Rodent-Borne Zoonotic Pathogens in Gedu, Bhutan
Article information
Abstract
Rodents are well-known reservoirs and vectors of many emerging and re-emerging infectious diseases, but little is known about their role in zoonotic disease transmission in Bhutan. In this study, a cross-sectional investigation of zoonotic disease pathogens in rodents was performed in Chukha district, Bhutan, where a high incidence of scrub typhus and cases of acute undifferentiated febrile illness had been reported in people during the preceding 4–6 months. Twelve rodents were trapped alive using wire-mesh traps. Following euthanasia, liver and kidney tissues were removed and tested using PCR for Orientia tsutsugamushi and other bacterial and rickettsial pathogens causing bartonellosis, borreliosis, human monocytic ehrlichiosis, human granulocytic anaplasmosis, leptospirosis, and rickettsiosis. A phylogenetic analysis was performed on all rodent species captured and pathogens detected. Four out of the 12 rodents (33.3%) tested positive by PCR for zoonotic pathogens. Anaplasma phagocytophilum, Bartonella grahamii, and B. queenslandensis were identified for the first time in Bhutan. Leptospira interrogans was also detected for the first time from rodents in Bhutan. The findings demonstrate the presence of these zoonotic pathogens in rodents in Bhutan, which may pose a risk of disease transmission to humans.
Bhutan is a small land-locked country known for its rich biodiversity and strong environmental and cultural conservation policies. The 2008 Constitution of the Kingdom of Bhutan decrees that the country will maintain a minimum of 60% of the total land under forest cover for all time [1], and currently 71% of Bhutan is under forest cover [2]. Bhutan has more than 200 species of mammals, including approximately 80 rodent species [3]. Rodents are well-known reservoirs and vectors of many emerging and re-emerging infectious diseases [4], but little is known about their role in zoonotic disease transmission in Bhutan. The number of acute undifferentiated febrile illness (AUFI) cases in Bhutan has been on the rise in recent years, and there have been several reports of scrub typhus outbreaks [5].
In this study, a cross-sectional survey for evidence of infection with Orientia tsutsugamushi and other zoonotic disease pathogens was conducted in rodents, in association with a case-control study, to investigate risk factors for scrub typhus in people who had AUFI in Bhutan. The rodent survey was conducted between February and April 2016 in 8 hamlets around Gedu (26°55′21.4″ N, 89°31′25.7″ E), a small town in Chukha district, south-western Bhutan, where a high incidence of scrub typhus had been detected in people in the case control study. Past studies have shown that the greatest risk of rodent-borne diseases is associated with anthropogenically disturbed habitats [6]. The constant change in agricultural practices around Gedu may thus be contributing to an increased risk of rodent-borne diseases in this area.
Rodents were collected alive using wire-mesh traps laid inside and near the vicinity of houses and vegetable gardens of 12 randomly selected households which were recruited as “cases” (from which scrub typhus had been confirmed in one or more household members) during the case-control study conducted in Gedu between October and December 2015. Traps were baited with processed cheese, meat, and dried fish and inspected daily during the study period. Captured rodents were transported to the laboratory where they were euthanized in accordance with a published protocol [7]. Rodent species were initially identified using morphological criteria from the South East Asian Murines Field Guide [8]. Immediately after euthanasia, kidney and liver samples were collected and preserved in 70% alcohol. Preserved tissue samples were transported to the Armed Forces Research Institute for Medical Sciences (AFRIMS), Bangkok, Thailand, for molecular identification of rodent species and phylogenetic analysis of zoonotic bacterial and rickettsial pathogens.
DNA was extracted from kidney and liver tissues using the Wizard® Genomic DNA purification kit (Promega, Madison, Wisconsin, USA) following a previously published protocol [9]. To perform rodent barcoding, the cytochrome c oxidase 1 gene (COI) was amplified from DNA extracts of rodent tissue using specific primers (BatL5310: 5′ CCTACTCRGCCATTTTACCTATG 3′, R6036R: 5′ ACTTCTGGGTGTCCAAAGAATCA 3′) following the protocol published by Herbreteau et al. [7]. PCR amplicons (648 bp) were sequenced and species were identified on the basis of percent DNA sequence identity with reference species deposited in the GenBank database, the Barcode of Life Data system (BOLD), or CERoPath project [7]. Amplification of species-specific genes was performed using published primers for each pathogen. Amplification products were purified using the QIAquick PCR Purification Kit (QIAGEN Inc., Valencia, California, USA) following the manufacturer’s instruction and sent for sequencing at AITbiotech Pte. Ltd. (Singapore). Sequence data were assembled using Sequencher 5.1 software (Gene Code Corporation, Ann Arbor, Michigan, USA) and consensus sequences were used for analyses. Sequences were aligned and constructed by identity matrix with the reference sequences using the Muscle algorithm implemented in Molecular Evolutionary Genetics Analysis (MEGA) 6.0 software. Maximum likelihood and Neighborjoining trees were constructed using MEGA 6.0 software as described by Tamura et al. [10], and bootstrap analyses with 1,000 resamplings were performed to test the robustness of the branching.
For pathogen detection, DNA extracts were screened for the presence of the rodent-borne pathogens causing leptospirosis, bartonellosis, rickettsiosis, scrub typhus, borreliosis, human granulocytic anaplasmosis, and human monocytic ehrlichiosis. The sequence of primers, probes, and conditions used for real-time and conventional PCR used in this study have been previously published, viz: Leptospira spp. lipL32 and 16S rRNA genes [11,12], Bartonella spp. ssrA and gltA genes [9,13,14], Rickettsia spp. 17 kDa [15], O. tsutsugamushi 47 kDa [16], Borrelia spp. 16S rRNA gene [17], Anaplasma phagocytophilum ankA gene and Ehrlichia chaffeensis TRP120 gene [18]. Positive samples were further characterized to species using DNA sequencing of pathogen-specific genes.
Twelve rodents were captured from a total of 176 trap nights across the 8 hamlets around Gedu. Serum, liver and kidney tissue samples were preserved from all 12 rodents and sent to AFRIMS for testing. Five species and 4 genera of rodents were identified using the COI gene sequence (Table 1; Fig. 1). Rodents were assigned to 4 species with sequence identities ranging from 95.0% to 100%. A phylogenetic tree was constructed using the Maximum Likelihood (ML) method based on the General Time Reversible model (GTR+G+I). The species identified were: the Himalayan field rat, Rattus nitidus (n=4, 99.8% identity); Chestnut white bellied rat, Niviventer fulvescens (n=1, 95.0% identity); house mouse, Mus musculus (n=1, 100% identity); and Asian house shrew, Suncus murinus (n=4, 96.4–96.6% identity). The remaining 2 rodents fell in the cluster of the Mus group but had a much lower sequence identity (89.6%) for which the closest reference sequence was for Mus pahari, Gairdner’s shrewmouse. These 2 rodents represent potentially novel species but more advanced morphological and molecular characterization is needed to verify this. All 4 of the S. murinus shrews were collected from a single household in Damdara, while R. nitidus rats were collected from 5 of the 8 collection sites.

Phylogenetic tree of COI gene (541 bp) of rodents sampled in the study. GenBank accession numbers, sequences from CERoPath project and BOLD are noted after each sequence. Only bootstrap values of 70% or greater are shown. Scale bar represents substitutions per site.
Pathogens were detected in 33.3% (4/12) of rodents (Table 1; Fig. 2A, B). A. phagocytophilum was detected from 16.7% (2/12) of rodent samples and was found in N. fulvescens and S. murinus. The pathogens B. grahamii and L. interrogans were each detected from a single R. nitidus (8.3%, 1/12). Two rodents, 1 N. fulvescens and 1 S. murinus, were co-infected with both Bartonella spp. and A. phagocytophilum. All rodent samples tested negative for Rickettsia, Borrelia spp., O. tsutsugamushi, and E. chaffeensis.
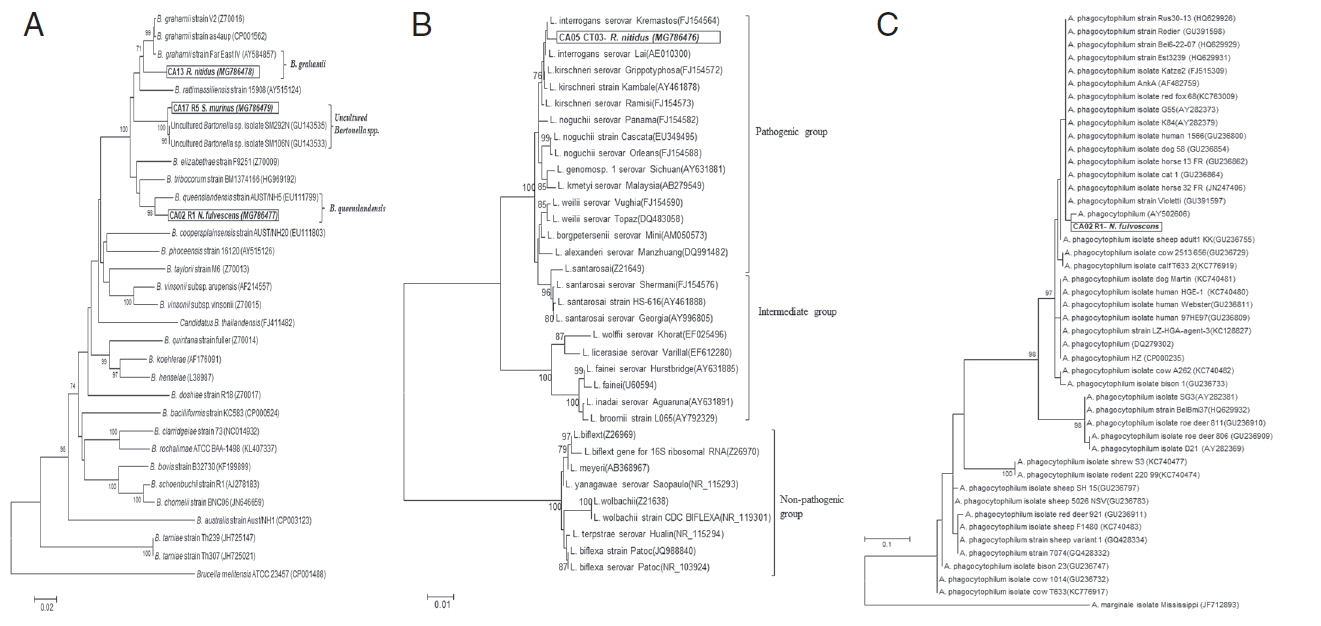
Phylogenetic relationship between pathogens detected in this study and reference sequences retrieved from GenBank database. (A) Neighbor-joining (NJ) tree constructed from gltA gene (655 bp) of Bartonella spp. using p-distance model. (B) Phylogenetic tree of 16S rRNA gene (1,169 bp) of Leptospira spp. constructed by NJ method based on the Maximum Composite Likelihood model. (C) Phylogenetic tree of ankA gene (50–101 bp) of Anaplasma spp. constructed by ML method based on the Kimura 2-parameter model. Only bootstrap values of 70% or greater are shown. Scale bar represents substitutions per site.
A phylogenetic analysis of the gltA gene sequence of Bartonella spp. (3 samples) and the 16S rRNA gene sequence of Leptospira spp. (1 sample) showed the highest sequence identity to B. queenslandensis (97.2%; GenBank accession no. MG786477), B. grahamii (96.9%; GenBank accession no. MG786478), uncultured Bartonella spp. (99.5%; GenBank accession no. MG786479) and L. interrogans (99.2%; GenBank accession no. MG786476); respectively (Fig. 2A, B). The ankA gene of Anaplasma spp. could not be amplified from 2 rodent samples. However, the DNA sequence of real-time PCR products (50–101 bp) showed the highest sequence identity (98.8%) to A. phagocytophilum strain red fox 51 (KC763008) using the BLASTN search program.
This study is the first report of A. phagocytophilum, B. grahamii, and B. queenslandensis in Bhutan. It is also the first report of L. interrogans in rodents from Bhutan, a pathogen that has previously been detected in cattle and human populations [19]. While only 12 rodents were captured and tested in this study, 33.3% were positive for the presence of the zoonotic pathogens investigated. It is therefore possible that rodents are commonly infected with zoonotic pathogens in Bhutan and play a significant role in disease transmission.
It is worth noting that all rodents were captured in and around the houses of confirmed cases of scrub typhus in humans that had been identified during the previous 4–6 months; however, none of the rodents tested positive for O. tsutsugamushi. The inability to detect the scrub typhus pathogen in the trapped rodents may be due to a number of factors, including the small sample size, trap locations and/or the time of year when the rodents were trapped. The incidence of scrub typhus in humans is highly seasonal and is typically highest from July to October, and lowest from February to May, in Bhutan. The rodents were trapped between February and April in this study, when there is a low incidence of scrub typhus in people, which may have been associated with a low prevalence of rodents infected with O. tsutsugamushi. Furthermore, traps may not have been placed in the environment where rodents serving as hosts for infected chiggers are likely to be found. All of our traps were laid inside and in the vicinity of houses, whereas a study conducted in Thailand found that the location of seropositive rodents was significantly associated with forest cover [20].
This study shows that sampling even a few rodents in Bhutan can provide important information about potential risks of rodent-borne zoonotic diseases. More detailed information on the geographic distribution and prevalence of these organisms in rodents could support the development of a list of potential pathogenic organisms for clinicians to test for in cases of undiagnosed undifferentiated acute fever. We recommend future studies are designed to capture a larger number of rodents across a wider range of localities and habitat types to obtain more accurate estimates of the prevalence of the zoonotic pathogens in rodents and association with different types of habitat. In future studies, the ectoparasites of rodents should also be collected and analysed. A One Health approach involving the contemporaneous collection and testing of samples from both humans and rodents in the study area is likely to contribute to a better understanding of the risk of rodentborne zoonotic diseases in Bhutan.
ACKNOWLEDGMENTS
We thank Dr. Kinzang Dukpa, Program Director of National Centre for Animal Health, Serbithang and Dr. Basant Sharma, Regional Director of Regional Livestock Development Centre, Tsimasham for providing technical and logistical support. The work has been managed by Massey University with financial assistance from the European Union under the One Health Program in Asia (EuropeAid/133708/C/ACT/Multi; Contract DCIASIE/ 2013/331-217). Laboratory testing of specimens was funded by the Armed Forces Health Surveillance Branch, Global Emerging Infections Surveillance and Response System (AFHSBGEIS), Silver Spring, Maryland, USA. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defence.
Notes
CONFLICT OF INTEREST
There is no conflict of interest related to this work.
