Genetic Diversity of Plasmodium vivax Causing Epidemic Malaria in the Republic of Korea
Article information
Abstract
Plasmodium vivax is more challenging to control and eliminate than P. falciparum due to its more asymptomatic infections with low parasite densities making diagnosis more difficult, in addition to its unique biological characteristics. The potential re-introduction of incidence cases, either through borders or via human migrations, is another major hurdle to sustained control and elimination. The Republic of Korea has experienced re-emergence of vivax malaria in 1993 but is one of the 32 malaria-eliminating countries to-date. Despite achieving successful nationwide control and elimination of vivax malaria, the evolutionary characteristics of vivax malaria isolates in the Republic of Korea have not been fully understood. In this review, we present an overview of the genetic variability of such isolates to increase understanding of the epidemiology, diversity, and dynamics of vivax populations in the Republic of Korea.
INTRODUCTION
The Republic of Korea has recently made significant progress in eliminating the burden of vivax malaria since the re-emergence of infectious vivax malaria in 1993, and has been categorized as one of 32 malaria-eliminating countries [1]. Malaria is an infectious disease caused by the Plasmodium parasite which is transmitted via the bite of infected female anopheline mosquitoes. Of the 4 Plasmodium species comprising P. falciparum, P. vivax, P. malariae, and P. ovale which routinely cause malaria in humans, P. vivax, a relatively neglected human malaria parasite, is the most widely distributed species in humans across the world causing an estimated 80–90 million cases each year [2]. Among the protozoa that cause malaria, P. falciparum is the greatest menace because of its high rate of mortality, mostly among children under the age of 5 in Africa. However, malaria caused by P. vivax is less lethal but has accounted for an enormous share of global malaria burden in recent years along with P. falciparum, greatly affecting the quality of life of several human populations in tropical, subtropical, and temperate countries [3,4]. P. vivax has evolved into a strain group with highly unique biological features, including a hypnozoite stage as a dormant and relapsing stage in the liver and a preference for invading reticulocytes (Duffy red blood cells) in as little as 0.5–1.5% normal blood; the lack of a convenient and continuous in vitro culture system for blood stages is an additional challenge [5]. In general, vivax malaria was thought to be benign and self-limiting, was not considered a priority in comparison with the significant burden of morbidity and mortality presented by P. falciparum, and was found to be harder to eliminate than P. falciparum [1,6,7]. However, the most geographically widespread distribution of P. vivax and recent evidence of severe and complicated vivax malaria across several endemic regions of the world suggest that this disease may have been overlooked as benign [8].
In the Republic of Korea, P. vivax is the most prevalent malaria species [9]. Vivax malaria has been endemic in the Republic of Korea for more than 2 decades after re-emergence. Since its re-emergence in 1993 [10], it has accounted for a total of 33,972 cases reported from 1993 to 2017 [9]. Ascertaining the P. vivax population structure is essential prior to establishing goals for control, elimination, and the rollout of interventions, and detailed knowledge of the spatial distribution, transmission, and clinical burden of P. vivax is required to act as a benchmark against which control targets can be set and measured [11–13]. The majority of studies on the genetic diversity of Plasmodium spp. have been based on P. falciparum, which is responsible for the most severe disease form, and on genes coding for antigenic determinants such as the circumsporozoite surface protein (CSP) and the merozoite surface protein (MSP). These antigenic genes are non-synonymous nucleotide polymorphisms and the multiple allelic forms differ in their ability to abrogate recognition by the host’s immune response [14]. At the time of re-emergence, no heterogeneity was observed in malaria incidence or in the distribution of P. vivax cases across the Republic of Korea. In July 1993, a Korean Army soldier stationed near the DMZ in northern Gyeonggi Province was diagnosed with vivax malaria [15]. After this first re-emergence case, there was an exponential increase in vivax malaria cases with epidemic outbreaks from 1995 to 2000. Since then, vivax malaria has been recognized as a significant public health problem [16–18]. In this short review article, we will discuss the characteristics of the well-known polymorphisms in the genes from South Korean isolates of P. vivax. The genetic diversity of the Plasmodium spp. population can be a valuable resource to analyze parasite populations. The degree of genetic diversity and the distribution of that diversity, or its population structure, offer insight into drifts in parasite transmission and parasite population history, information that is pivotal to malaria control and elimination programs and assessing the impact of such interventions [19].
GENETIC DIVERSITY OF WELLCHARACTERIZED POLYMORPHIC REGIONS IN P. VIVAX ISOLATES FROM THE REPUBLIC OF KOREA
Because the high levels of polymorphism in Plasmodium parasites indicate the sophisticated immune evasion strategies of malaria parasites [20], understanding parasite population diversity is important for the strategic development and deployment of control measures [21,24]. Moreover, genetic diversity based on multiple gene polymorphisms in vivax parasites has been shown to greatly contribute to drug resistance [25]. Due to polyclonal infection of the same host by multiple parasite clones, to elucidate local population structure and genetic diversity, it is essential to be able to distinguish between distinct clones as well as between infections. In addition, P. vivax in the Republic of Korea displays relatively low parasitemia and low male-to-female gametocyte ratio, contributing to low transmission efficacy [26,27]. Further, the frequency of the polyclonal infection rate in the Republic of Korea is relatively very low (~3% vs 30–65% in Papua New Guinea, Thailand, and India) [28]. These unique biological characteristics are not well-understood. In fact, it is well-known that P. vivax displays much greater genetic diversity in terms of SNP diversity and microsatellite and gene family variability than P. falciparum [29], and this means that the global population of P. vivax may have a capacity for greater functional variation [29]. Understanding the genetic variation patterns of the parasite may help in analyzing trends and assessing the extent of malaria endemicity in the Republic of Korea [30]. This knowledge of the extent of correlation between the parasite population diversity and transmission intensity could also provide early warnings to identify populations living in endemic areas, facilitate the identification of new drug resistance genes, and eventually provide information essential for malaria control and elimination [31]. The well-characterized polymorphic regions in genes such as gam1 coding for the gametocyte antigen 1, csp coding for the circumsporozoite protein (CSP), msp1 and msp3α coding for the merozoite surface proteins (MSP)-1 and 3α, respectively, ama1 coding for the apical membrane antigen (AMA) 1, and dbp coding for the Duffy binding protein (DBP) have been extensively used to analyze genetic diversity patterns of natural populations of P. vivax (Table 1) [32,33].
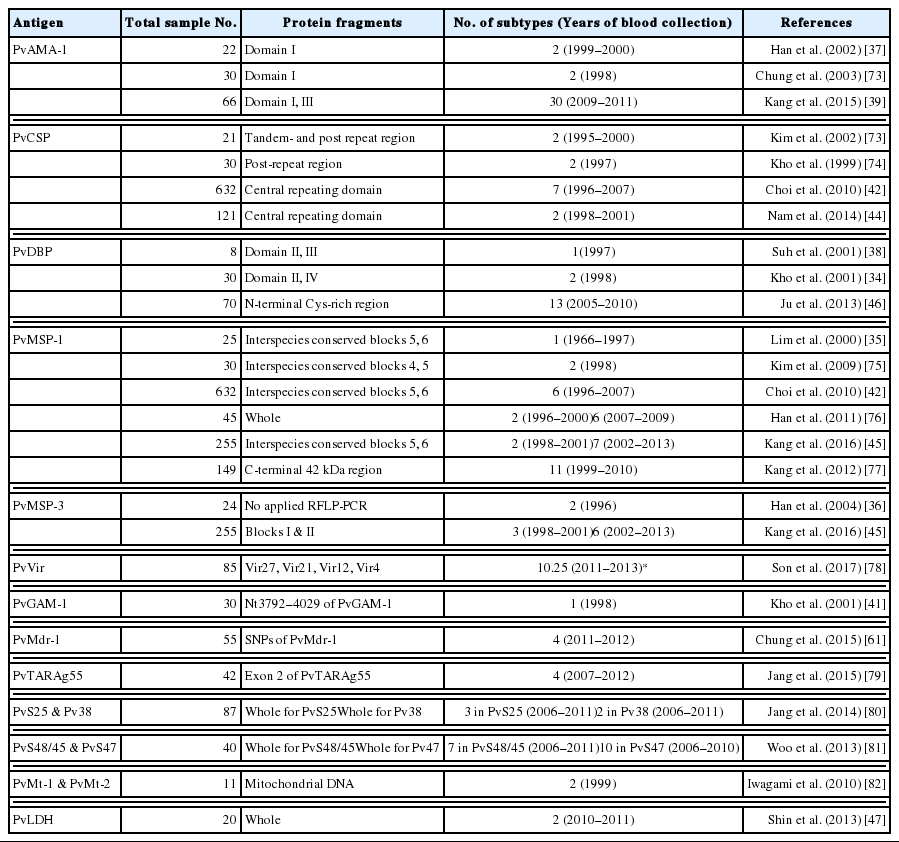
Genes encoding antigens used as molecular markers to study field populations of Plasmodium vivax in the Republic of Korea
The genetic diversity of South Korean P. vivax isolates has been extensively studied and characterized by several research groups. The population structure of P. vivax parasites in the Republic of Korea has been largely based on genetic analyses focusing on the mutation of antigen-coding genes. Initially, a combinational analysis of PvMSP-1 and PvMSP-3α clearly demonstrated the drastic genetic changes occurring in the South Korean P. vivax population. The MSP-1 and 3α are blood-stage vaccine-candidate antigens with repetitive arrays. In particular, PvMSP-3α is highly polymorphic and has been confirmed to be a molecular marker in several geographic malaria parasite populations [32]. From early re-emergence to 2000, PvDBP, PvMSP-1, and PvMSP-3α genotypes were found to be highly homogenous. However, since 2001, they have rapidly diversified. Based on the investigation of 30 samples from P. vivax patients in 1998, it was found that PvDBP had little polymorphism and only 2 genotypes of P. vivax coexisted in the Republic of Korea [34]. PvDBP is involved in the formation of an irreversible junction between P. vivax merozoites and its receptor, the Duffy antigen/receptor for chemokines at the surface of immature reticulocytes, and is a prime blood stage vaccine candidate antigen for P. vivax. PvMSP-1 in South Korean isolates analyzed from blood samples collected in 1996 and 1997 was reported to be highly homogenous [35]. The PvMSP-3α locus of re-emerging Korean P. vivax is dimorphic with 2 allelic types coexisting in the endemic area [36]. In addition, 2 genotypes of PvAMA-1 genes coexist in the reemerging South Korean P. vivax [37], suggesting that 2 genotypes of P. vivax coexist in the endemic area of the Republic of Korea. PvAMA-1 is an immunogenic, type I integral membrane protein, which is expressed at the apical surface of merozoites and sporozoites. Because of the geographical linkage of the genetic variation of these genes in Plasmodium spp, P. vivax found in the Republic of Korea was unique and, based on molecular structure analysis, could be divided into 2 strains [38]. Of course, the molecular basis for the geographic linkage is poorly understood to-date. Based on the analysis of nucleotide diversity across the entire PvAMA-1 gene, South Korean PvAMA-1 as found to have limited diversity compared to global isolates and differed from other global isolates in its pattern of genetic polymorphism [39]. In case of polymorphism in GAM-1, a P. vivax transmission-blocking candidate antigen [40] no longer used as a reliable genetic marker [32], Korean isolates from 30 patients in 1998 showed only one genotype containing 4 33-bp motifs identical to the Belem strain [41]. However, an analysis of South Korean isolates from 1996–2007 based on the sequence analysis of PvMSP-1 and PvCSP genes revealed that most isolates after 2000 were different from the isolates during the initial re-emergence period [42]. The PvCSP is an abundant antigen on the surface of sporozoites, which is extensively used as a vaccine development target [43]. In the above study, 6 PvMSP-1 subtypes were identified in genomic parasite DNA from 632 patients and a new PvCSP subtype was found based on polymorphism of repeat regions. These authors also found that malaria parasites in the Republic of Korea were becoming progressively more genetically diverse. Interestingly, when PvCSP was loaded onto an RDT kit and applied with 121 patient sera, CSP antigen-antibody reactions were seen in up to 57% of cases [44]. Similarly, Kang et al. [45] suggested that the P. vivax population in the Republic of Korea has been diversifying rapidly and that new genotypes are appearing, based on combinational genetic analysis of polymorphic patterns of 2 major antigenic proteins, PvMSP-1 and PvMSP-3α. They also found that the PvDBP II in Korean isolates had a polymorphic nature and that 13 different PvDBP II haplotypes were present in Korean P. vivax isolates. This population is under natural selection, and PvDBP II showed limited genetic polymorphism at restricted positions with a lower genetic diversity [46]. However, PvLDH in Korean isolates, a useful antigen for a rapid diagnostic test (RDT), did not show any genetic variation except a single-nucleotide polymorphism with no effect on the amino acid sequence [47].
In summary, genetic analyses using antigen-coding genes such as PvDBP, PvCSP, PvMSP (MSP3α and MSP3β), PvAMA, and PvGAM-1 suggest that only 1 or 2 alleles existed in South Korean isolates before 2000. However, these antigens have shown their patterns by year genetic diversity since then. It is not certain why the South Korean P. vivax population structure has changed so drastically since the early 2000s despite rapid reductions in transmission and low multiple infection rates. The rarity of polyclonal P. vivax infection in the Republic of Korea [48] suggests that local outcrossing alone is unlikely to explain the increased genetic divergence observed in the antigen-coding genes. Thus, cases introduced from an external reservoir likely contributed to the increased diversity [28]. Because changing antigenicity is a major strategy for parasites to be protected from host immunity, antigenic diversity/polymorphism, which links the expression of different alleles of a gene in different parasite populations with outcomes of selection under immune pressure, would selectively expand [49].
A SPECTRUM OF POPULATION STRUCTURES IDENTIFIED VIA MICROSATELLITE MARKER ANALYSIS
P. vivax populations are more genetically diverse and more frequently comprise polyclonal infections than do sympatric P. falciparum, suggesting that frequent recombination between the clones in the populations may contribute to genetic diversity [50]. Although P. vivax surface antigens show high genetic diversity, antigen-encoding genes can be subject to strong natural selection due to the host immune system. In contrast, microsatellites (MSs) are putatively neutral markers that can be used to monitor parasite transmission patterns without the potentially confounding impact of host immunity or other selective pressures. Simple MSs (sequence tandem repeats) are a valuable resource for the study of genetic diversity. They are useful for fingerprinting an organism because they are genetically hypervariable, co-dominant and locus-specific, and are not directly subject to host immunity, making them compatible with an important assumption of population genetic theory which states that molecular markers should be selectively neutral [51]. In case of P. falciparum, microsatellite markers have been known to be extensively widespread occurring every 2–3 kb throughout the genome [52]. In spite of a limited number of polymorphic MS loci in P. vivax (a few dozens) rather than that in P. falciparum (~1,000), MS-based studies of P. vivax have provided pivotal information regarding population structure and diversity [32,53,54]. The recent markers of choice for large-scale population studies are highly polymorphic and short (1–6 bp-long) MSs [55]. P. vivax contains 14 chromosomes. There are now 5 reference genomes of P. vivax (Salvador I, North Korean, India VII, Mauritania I, and Brazil I) [29] and P. vivax chromosomes appear to significantly larger and have more stable GC-rich genomes (approximately 40%) than those of P. falciparum (19%) [56,57]. Using 13 MS DNA markers across 12 chromosomes in P. vivax from the Republic of Korea, only 2 major multilocus MS genotypes were found during 1997–2000 while numerous genotypes appeared in 2007, indicating a marked expansion in the genome-wide genetic diversity of the P. vivax population structure in the Republic of Korea [26,51]. In addition, Iwagami et al. [58,59] investigated P. vivax in South Korean soldiers using 14 highly polymorphic MS DNA loci and dividing the samples from soldiers and veterans into 3 different time periods spanning 1993–1998, 1999–2003, and 2004–2008 in a 15-year-long longitudinal study on P. vivax population genetics. Further, another population genetic approach using P. vivax positive blood samples from 98 autochthonous cases and nine tandem repeat markers showed a trend of increasing parasite divergence since re-emergence [48]. It is also reported that polyclonal infections were rare (~3%) suggesting that local out-crossing alone was unlikely to explain the high diversity. The South Korean Army introduced a strategy of mass chemoprophylaxis with chloroquine during the malaria transmission season and primaquine after the transmission season to reduce the number of vivax malaria cases in both the military and civilian populations. Chemoprophylaxis with chloroquine (300 mg/wk) and primaquine (30 mg/wk) began with approximately 16,000 soldiers in 1997 and was expanded to more than 200,000 military personnel in zones at high risk for vivax malaria [60]. In a process of monitoring for chloroquine-resistant P. vivax using single nucleotide polymorphisms in Pvmdr1 and microsatellite genotypes, P. vivax with newly identified SNPs in Pvmdr1 was introduced and the microsatellite-based study revealed higher genetic diversity in samples collected in 2012 than in those collected in 2011 [61]. The study determined microsatellite genotypes of the 55 isolates based on a combination of allelic data of 10 microsatellite loci. The findings revealed that 2 dominant MS haplotypes had been transmitted for several years, providing decisive evidence for a low recombination rate. This may be due to recent migrations of different genotypes from other geographic regions to the Republic of Korea, and suggests an increase in the genetic diversity of the P. vivax population in the Republic of Korea [45,48]. While the level of genetic diversity in the P. vivax population in the Republic of Korea is significantly lower than that observed in tropical and subtropical areas [62–65], the population diversity is consistently increasing and is associated with a decline in multi-locus linkage dis-equilibrium (LD) over time [28,48,51,59]. Declining LD may also result from inter-allelic recombination. For example, PvMSP-1 displays heterogeneity in its frequency of inter-allelic recombination events, leading to rapid decline in multilocus LD between pairs of synonymous sites [66]. This is to be possible that the appearance of a P. vivax parasite population with new genotypes from other external reservoir(s) has accelerated genetic diversity in the South Korean P. vivax population through recombination events. These facts might be consistent with the low polyclonal infections and the small numbers of imported malaria cases in the Republic of Korea (<20 cases per year except 52 in 2000) [9].
CONCLUSION
Based on the above-mentioned studies on genetic diversity and incidence, the evidence obtained supports a link between vivax malaria in South and North Korea [67]. Annual WHO Malaria Reports show that North Korea had a restricted capacity to eliminate malaria until 1998 and experienced its highest reported incidence of malaria in 2001, peaking at 296,540 microscopically confirmed and indigenous cases (43.4 cases per 1,000 population) [68]. The vivax malaria cases in North Korea coincided with peak incidence in South Korea (3,932 in 1998, 3,621 in 1999, 4,142 in 2000, and 2,556 in 2001) [9]. Several independent genetic studies have reached the conclusion that the extant P. vivax population is shaped by continuous introductions from an external source, possibly North Korea, although the exact genetic data on North Korean vivax populations is not available to-date. This scenario does not exclude the consideration that local transmission events may also occur in South Korea and contribute to the population structure. Importantly, this interpretation allows for local propagation to occur largely via inbreeding, which is concordant with the low incidence, low prevalence of infections, and limited seasonality (July-August) of the opportunity for recombination in the An. sinensis vector. This scenario also supports the longitudinal trends in the genetic diversity and structure of P. vivax in the Republic of Korea. Indeed, there is an example elsewhere similar to such genetic diversity in the Republic of Korea. Sri Lanka has remarkably reduced its vivax malaria incidence case by 99.99% with high levels of heterozygosity of P. vivax and large effective population sizes [69]. Moreover, local vivax populations are characterized by high levels of allelic polymorphism and significant LD indicative of reduced recombination. However, approximately 70% isolates in that study had polyclonal infections. The authors suggested that the imported malaria could by the most likely source of vivax malaria transmission. Overall, the significant diversity of the P. vivax population observed hampers the development of an effective vaccine against South Korean isolates of vivax malaria. In comparison to the very low endemic, pre-elimination settings of Sabah, Malaysia [70] and Southern Iran [71], there is lesser evidence of population bottlenecking in the Republic of Korea.
Therefore, further continuous monitoring of malaria parasites using various molecular markers including polymorphic DNA markers in the P. vivax genome is needed for the control and elimination of vivax malaria in the Republic of Korea. Such monitoring can also assess the population structure and dynamics of the parasite transmission.
ACKNOWLEDGMENTS
This research was supported by the grants of a Research Grant from Inha University (56866-01, TSK).
Notes
CONFLICTS OF INTERESTS
The authors declare they have no competing interests.