Molecular Detection and Phylogenetic Analysis of Anaplasma phagocytophilum in Horses in Korea
Article information
Abstract
The identification and characterization of pathogenic and zoonotic tick-borne diseases like granulocytic anaplasmosis are essential for developing effective control programs. The differential diagnosis of pathogenic Anaplasma phagocytophilum and non-pathogenic A. phagocytophilum-like Anaplasma spp. is important for implementing effective treatment from control programs. The objective of the present study was to investigate the prevalence of Anaplasma spp. in horses in Korea by nucleotide sequencing and restriction enzyme fragment length polymorphism assay. Of the 627 horses included in the study, only 1 (0.2%) was infected with A. phagocytophilum. Co-infection with A. phagocytophilumlike Anaplasma spp. was not detected in the study. The 16S rRNA sequence of A. phagocytophilum was similar (99.5–100%) to A. phagocytophilum 16S rRNA isolated from horses in other countries. PCR adapted to amplify A. phagocytophilum groEL and msp2 genes failed to generate amplicons, suggesting genetic diversity in these genes. This study is the first molecular detection of A. phagocytophilum in horses in Korea. Human granulocytic anaplasmosis and animal infection of A. phagocytophilum have been reported in Korea recently. Because of vector tick distribution, global warming, and the increase of the horse industry, horses should be considered as a potential reservoir for A. phagocytophilum, and cross infectivity should be evaluated even though a low prevalence of infection was detected in this study. Furthermore, continuous surveillance and effective control measures for A. phagocytophilum should be established to prevent disease distribution and possible transmission to humans.
INTRODUCTION
Anaplasma phagocytophilum is the causative agent of granulocytic anaplasmosis in humans, dogs, cats, and horses, and tick-borne fever in domestic ruminants [1]. In horses, equine granulocytic anaplasmosis (EGA) caused by A. phagocytophilum is characterized by thrombocytopenia [2]. A. phagocytophilum infection in humans causes human granulocytic anaplasmosis (HGA). Acute infections are self-limiting simultaneously with the increased adaptive immune responses and the activation of macrophages [3]. Granulocytic anaplasmosis is developed by the suppression of, or disorder in host defenses which pre-dispose horses to opportunistic and/or secondary infections with virus, bacteria, and fungi [4].
The rapid and exact diagnosis of zoonotic and pathogenic diseases such as HGA is important for risk assessment in tick-borne disease control programs [5]. Therefore, it is essential to differentiate between pathogenic A. phagocytophilum and A. phagocytophilum-like Anaplasma spp. (APL) that do not cause clinical symptoms in infected animals and are considered non-pathogenic [5].
To date, multiple species of Anaplasma have been detected in Korea by PCR – A. phagocytophilum in shelter dogs [6] and shelter cats [7]; A. phagocytophilum and A. bovis in Korean water deer [8]; A. bovis in Korean spotted deer [9]; A. phagocytophilum, A. bovis, and A. centrale in ticks [10–14]; and A. phagocytophilum and APL in cattle [15].
On the other hand, A. phagocytophilum in horses has been only detected by immunofluorescent antibody (IFA) test [16] and by ELISA [17]. Although 16 samples were seropositive in the previous study [17], none of the samples tested positive for A. phagocytophilum 16S rRNA by nested PCR (nPCR). This indicates that the horses were previously exposed. In this direction, we assessed the prevalence, risk factors, and co-infectivity of A. phagocytophilum and APL in horses in Korea by using nPCR.
MATERIALS AND METHODS
Ethics statement
This study conducted during 2016–2017, failed to receive approval from the Institutional Animal Care and Use Committee (IACUC) at Kyungpook National University (KNU), as IACUC at KNU evaluates laboratory animals maintained in indoor facilities and does not regulate research involving outdoor animals. Blood samples were collected by practicing veterinarians at local clinics during monitoring, surveillance, and treatment, or during regular check-ups after receiving verbal consents from the farm owners.
Sample size determination and sample collection
In 2016, the total number of horses reared in Korea was recorded as 27,676 [18]. The sample size was determined using the following formula with an expected disease prevalence of 10%, an accepted absolute error of 5%, and a confidence level of 95% with a simple random sampling design [19]:
where n=required sample size, pexp=expected prevalence, and d=desired absolute precision.
According to the formula, a minimum of 138 samples was required, but 627 horses were randomly selected from the whole country using a simple random sampling method during monitoring or surveillance in 2016–2017 (Fig. 1). Whole blood samples were collected and were stored at −20°C until use. Data on age, sex, breed, and region were recorded for analysis.
nPCR
Genomic DNA was extracted from the whole blood sample, using a commercial DNeasy Blood and Tissue kit (Qiagen, Melbourne, Australia) following the manufacturer’s instructions. The extracted DNA was stored at −20°C until use. The AccuPower HotStart PCR Premix kit (Bioneer, Daejeon, Korea) was used for PCR amplification. nPCR amplification of 16S RNA using 2 primer pairs EE1/EE2 and EE3/EE4, with an expected amplicon of 928 bp was used to screen for Anaplasma spp. infection [20]. For species identification, PCR-positive samples were re-amplified to identify the 16S rRNA genes of A. phagocytophilum using the primer sets EE1/EE2 and SSAP2f/ SSAP2r [21,22], generating an expected amplicon of 641 bp. Gene fragments A. phagocytophilum heat shock protein (groEL) and major surface protein 2 (msp2) were also amplified using nPCR for the detection and/or characterization of A. phagocytophilum [6]. A. phagocytophilum detected from a dog [6] and A. phagocytophilum and APL detected from cattle [15] were included as positive controls.
Subtyping by restriction enzyme fragment length polymorphism (RFLP) assay
To subtype the strains and distinguish between A. phagocytophilum and APL, the partial sequences from 16S rRNA, which were amplified using primers SSAP2f/SSAP2r by nPCR, were used for RFLP assay [5]. XcmI and BsaI restriction enzymes (New England Biolabs, Hitchin, UK) were selected to discriminate between A. phagocytophilum and APL, and between APL clades A and B, respectively [5].
DNA sequencing and phylogenetic analysis
The 16S rRNA amplicons from the infected animal were purified using the QIAquick Gel Extraction Kit (Qiagen), ligated into pGEM-T Easy vector (Promega, Madison, Wisconsin, USA), and transformed into Escherichia coli DH5α-competent cells (Thermo Fisher Scientific, Wilmington, Delaware, USA) and then incubated at 37°C overnight. Plasmid DNA extraction was then performed using a plasmid miniprep kit (Qiagen) following the manufacturer’s instructions.
Recombinant clones were selected and sent to Macrogen (Seoul, Korea) for sequencing. The sequences were aligned and analyzed using the multiple sequence alignment program CLUSTAL Omega (v. 1.2.1), and the alignment was corrected using BioEdit (v. 7.2.5). Phylogenetic analysis was performed using MEGA (v. 6.0) based on the maximum likelihood method employing the Kimura 2-parameter distance model. The aligned sequences were analyzed using a similarity matrix. The stability of the trees obtained was estimated by bootstrap analysis for 1,000 replicates.
Statistical analysis
Chi-square test was performed to analyze significant differences among the groups and a value of P <0.05 was considered to indicate statistical significance. GraphPad Prism (v. 5.04; GraphPad Software Inc., La Jolla, California, USA) was used for statistical analyses. A 95% confidence interval (CI) was calculated for all estimates.
RESULTS
nPCR and RFLP
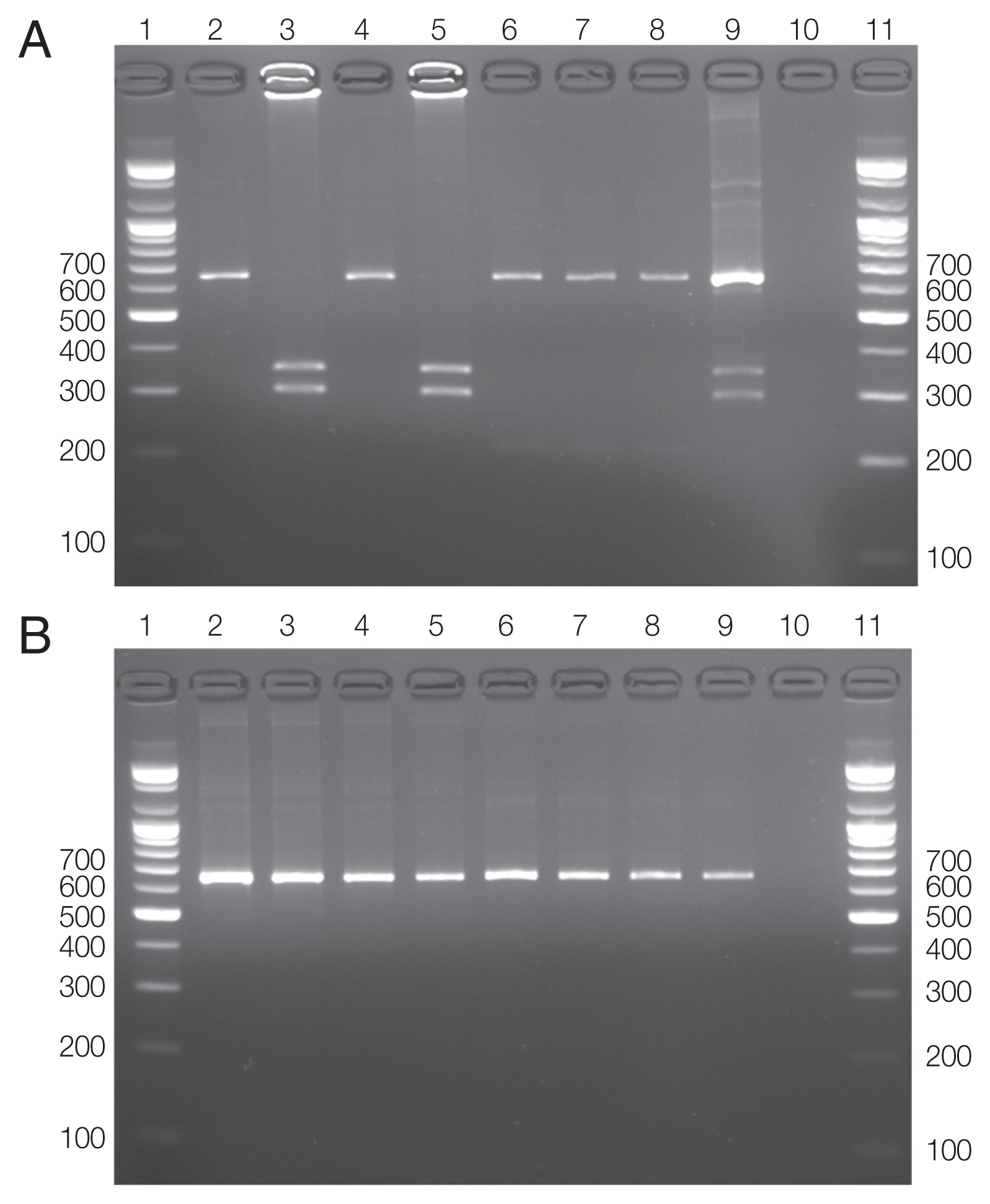
Of the 627 horses, nPCR amplification of 16S rRNA gene fragments using the EE3/EE4 and SSAP2f/SSAP2r primer pairs identified only 1 (0.2%) horse as positive for Anaplasma spp. (Table 1). None was positive for A. phagocytophilum groEL and msp2 gene fragments. The first discrimination between A. phagocytophilum and APL was accomplished by digesting the 16S rRNA amplicons (641–642 bp) with XcmI. A. phagocytophilum amplicons were cut to generate 344 and 297 bp fragments (Fig. 2A; lanes 3 and 5), while amplicons from APL were not cut by this enzyme (Fig. 2A; lane 7). In the second step, digestion of the same amplicons with BsaI distinguished APL clade A from clade B. APL clade B generated 2 fragments of 422 (or 423) and 219 bp, while the amplicon from APL clade A was not digested by this enzyme (Fig. 2B; lane 7). Co-infections of A. phagocytophilum and APL were detected (Fig. 2A; lane 9) by XcmI, and finally, APL was identified to belong to the APL clade A (Fig. 2B; lane 9) using BsaI. RFLP revealed that the infected horse carried A. phagocytophilum. Co-infection with APL was not detected in the infected animal. The horse positive for A. phagocytophilum was a 5-year old castrated thoroughbred in the southern region (Table 1). No statistically significant differences were observed with respect to any variables (P >0.05).

Prevalence of Anaplasma phagocytophilum using 16S rRNA gene in horse bloods in Korea during 2016–2017
Nucleotide sequencing and phylogenetic analysis
The results from the RFLP assay was confirmed by 16S rRNA sequencing. The A. phagocytophilum 16S rRNA sequence (HGN- 20) obtained in this study has been deposited to GenBank (accession number MH794246). Phylogenetic analysis showed that the 16S rRNA sequence of A. phagocytophilum (Fig. 3) clustered together with previously deposited A. phagocytophilum sequences.
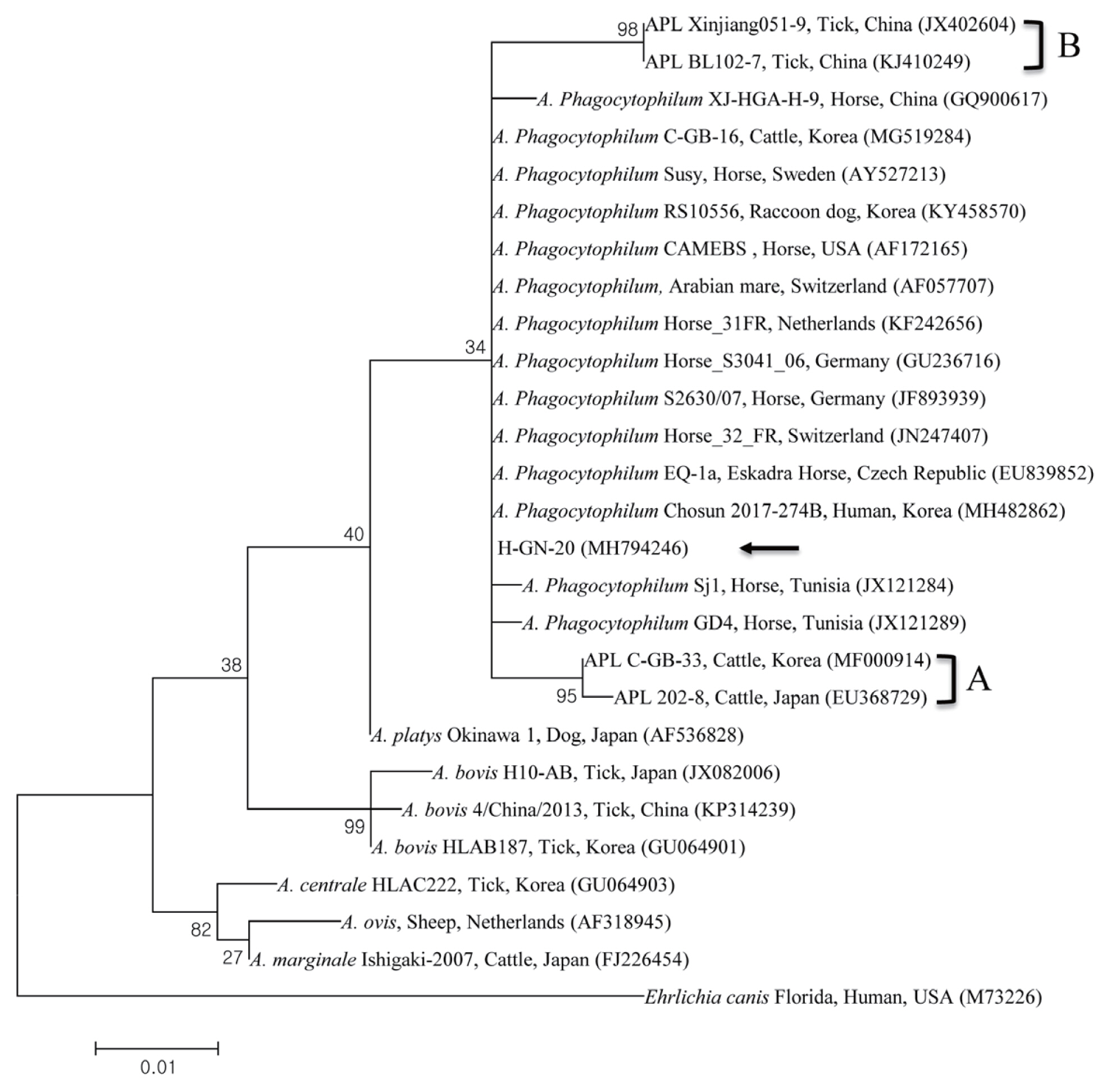
Phylogenetic tree constructed using maximum likelihood method, based on the 16S rRNA nucleotide sequences of Anaplasma phagocytophilum. The black arrow indicates the sequence analyzed in this study. A and B, two clades of APL.
The A. phagocytophilum 16S rRNA sequence shared 99.5–100% identity to several A. phagocytophilum sequences from Sweden, Switzerland, USA, and Korea, including the ones from cattle in Korea (100%, MG519284) [15], a horse in the USA (99.7%, AF172165), a horse in Sweden (99.5%, AY527213), and a horse in Switzerland (99.5%, AF057707).
DISCUSSION
Previous serological studies from Korea detected A. phagocytophilum in horses (2.2%, 2/92) reared on Jeju Island by IFA test [16] and A. phagocytophilum (2.9%, 16/549) in horses by ELISA between 2009 and 2013 [17]. Although molecular studies were carried out among 549 horses in the same study, A. phagocytophilum could not be detected by PCR [17]. Anaplasma infection in horses has been described in other Asian countries, including A. phagocytophilum (2.1%, 120/486) by ELISA in Taiwan [2], A. phagocytophilum (10.7%, 16/150) by PCR in Pakistan [23], and A. phagocytophilum (3.4%, 3/87) by IFA in Japan [24].
In this study, only 1 (0.2%) of the 627 horse blood samples tested positive for A. phagocytophilum by nPCR analysis for 16S rRNA gene fragments. Additionally, RFLP analysis also identified it as A. phagocytophilum. The horse positive for A. phagocytophilum was a 5-year old castrated thoroughbred from Gyeongnam province in the southern region. This horse was a riding horse at a local community located in the southernmost city of Korea. The prevalence of Anaplasma spp. may differ between geographic locations in association with tick habitat and distribution [21,25]. Korea is gradually changing to a subtropical climate zone due to global warming, and Gyeongnam province is located at a lower latitude which favors tick survival. This difference may clarify the observed differences in the prevalence of ticks and tick-borne diseases. A similar study reported that the horses in the southern region of Korea had significantly higher seropositivity than those in other regions [17]. Furthermore, molecular prevalence (0.2%) in the present study was lower than those of serological studies in Korea [16,17], Taiwan [2], and Japan [24] which have similar climate conditions. This might be attributed to the fact that antibodies persist in the blood longer than the antigen. Prevalence was different between breeds and a thoroughbred horse was found to be infected in the present study. Previous studies reported a higher seroprevalence in thoroughbred horses in Tunisia and warmblood horses in Korea than in other breeds [17,26].
In the present study, the infected horse did not present any clinical symptoms at the bleeding time. Generally, EGA presents with fever with ataxia, reluctance to move, distal limb edema, and depression. The symptoms of granulocytic anaplasmosis are nonspecific and this could describe why it was not diagnosed [27]. Clinical anaplasmosis in horses is perhaps underdiagnosed as most horses recover naturally and clinical symptoms are similar to those caused by infections with other pathogens such as equine herpes virus, Babesia caballi, Theileria equi, equine infectious anemia virus, Leptospira, Borrelia burgdorferi, and equine arteritis virus [28].
This is the first study on the molecular detection of A. phagocytophilum infection in horses in Korea. We observed a relatively lower prevalence of A. phagocytophilum infection compared to other studies in Korea – A. phagocytophilum (42/66, 63.6%) in Korean water deer [8]; A. phagocytophilum (20/764, 2.6%) and APL (16/764, 2.1%) in cattle [15]; and A. phagocytophilum (89/266 pools, 24.5%) in ticks collected from Korean water deer (Hydropotes inermis argyropus) [12]. This might be attributed to the differences in the study regions, hosts, breeds, vector ticks, sampling seasons, and climate. Other studies have reported a similar low prevalence in Korea – A. phagocytophilum (1/1,058, 0.1%) in shelter dogs [6], A. phagocytophilum (2/222, 0.9%) in shelter cats [7]. The prevalence of Anaplasma spp. were higher in wild animals and vector ticks as compared to domestic animals [6–8,12,15].
In Korea, the horse industry is gradually growing annually; the horse industry related indices for 2016 are higher than in 2015–2,278 horse industry companies, 479 horse-riding courses, and 16,662 horse-associated employees in 2016, whereas 2,052 horse industry companies, 457 horse-riding courses, and 15,845 horse-associated employees in 2015 [18]. Therefore, it is more important to prevent the spread of zoonotic diseases such as EGA between horses and humans. In this study, the infected horse was used for riding at a horse-riding institute, frequently came in contact with people, and was also allowed to graze freely in the nearby grassland area, thus allowing easy access to tick vectors. Seroprevalence is thoroughly related to the activity of horse – high seroprevalence was detected in brood mares and horses used in riding clubs [26].
The genetic diversity of genus Anaplasma has been described by analyzing nucleotide sequences from diverse genes [29]. Although we tried to amplify groEL and msp2 genes of A. phagocytophilum, PCR failed to amplify these genes, suggesting that their genetic diversities do not entirely explain the role of genotypes and ecotypes in epidemiological cycles [29]. While gro-EL and msp2 genes could differentially identify A. phagocytophilum, 16S rRNA sequencing showed high identity to sequences from Korea and other countries previously deposited to Gen-Bank; thus, 16S rRNA sequencing was enough to identify the species of Anaplasma in this study.
The pathogenic and zoonotic tick-borne diseases caused by Anaplasma spp. are crucial, so it is critical to distinguish between A. phagocytophilum and APL (clades A and B) [5]. The deficiency of clinical symptoms in infected animals and the unique tick vectors further suggest that this novel Anaplasma strain (APL clade A) primarily found in Japan [30] might possibly be a new species. While the Anaplasma spp. strains identified in sheep and ticks infesting cattle differed from APL clade A and from all other classified and unclassified Anaplasma strains (APL clade B) in China [31]. Therefore, we additionally performed an RFLP analysis to distinguish between A. phagocytophilum and APL infection. Our results revealed that there was no co-infection with APL. RFLP assays do not need sequencing and cloning to evaluate strain prevalence and co-infection. Furthermore, it decreases the time spent and the costs of molecular diagnosis [5].
In conclusion, this is the first study on the molecular detection of A. phagocytophilum in horses reared in Korea. We analyzed some risk factors for A. phagocytophilum in this study. Since the A. phagocytophilum isolates from humans clustered with those from horses and cattle, the prevalence of EGA would have public health implications, necessitating effective control of EGA. Equine veterinarians should consider differential diagnosis of anaplasmosis from other febrile diseases. Further studies are needed to assess the clinical relevance of A. phagocytophilum and APL. Although previous studies have led to a better understanding of the evolutionary behavior and host-pathogen relations of Anaplasma spp., additional studies are required to identify the reservoir and vectors of Anaplasma spp. in Korea.
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.