Anaplasma marginale and A. platys Characterized from Dairy and Indigenous Cattle and Dogs in Northern Vietnam
Article information
Abstract
Anaplasma marginale and A. platys were detected and characterized (16S rDNA sequence analysis) from dairy and indigenous cattle, and the latter in domestic dogs in Vietnam. A phylogenetic tree was inferred from 26 representative strains/species of Anaplasma spp. including 10 new sequences from Vietnam. Seven of our Vietnamese sequences fell into the clade of A. marginale and 3 into A. platys, with strong nodal support of 99 and 90%, respectively. Low genetic distances (0.2–0.4%) within each species supported the identification. Anaplasma platys is able to infect humans. Our discovery of this species in cattle and domestic dogs raises considerable concern about zoonotic transmission in Vietnam. Further systematic investigations are needed to gain data for Anaplasma spp. and members of Anaplasmataceae in animal hosts, vectors and humans across Vietnam.
Anaplasma species of the family Anaplasmataceae (order Rickettsiales) are intracellular bacteria, transmitted by ticks, which cause diseases (anaplasmosis) in domesticated ruminants, dogs, wild animals and humans worldwide [1,2]. Ticks of the genus Ixodes are responsible for zoonotic transmission of Anaplasma spp. to humans [3]. Seven species are common bovid hemopathogens, namely Anaplasma bovis, A. capra, A. centrale, A. marginale, A. ovis, A. phagocytophilum, and A. platys [2]. The intra-erythrocytic A. marginale is found globally as the most prevalent causative agent for bovine anaplasmosis, and the intra-leukocytic A. phagocytophilum causes anaplasmosis of a range of animals and humans [2,4–6]. Anaplasma platys is a common cause of leukopenia and neutropenia in dogs, ruminants, horses and humans [2,7–10]. Ruminants are the hosts for at least 5 Anaplasma species (A. marginale, A. centrale, A. bovis, A. phagocytophilum, and A. platys), and are a significant reservoir for human infections [5,6].
Molecular identification of Anaplasma spp. and other rickettsial pathogens, ie., Ehrlichia spp. and Neorickettsia spp. in ticks, animals and humans, has been frequently based on single 16S ribosomal DNA (16S rDNA or rrs), or in combination, with 60 kDa heat-shock protein gene (groEL; or HSP60), beta subunit of RNA polymerase (rpoB), citrate synthase (gltA), and major surface protein 4 gene (mps4) sequences [1,2,4,6,8,11–13]. The systematics of the family Anaplasmataceae has been completely revised with the assistance of molecular genetic markers [1–3].
In Vietnam, anaplasmosis has been investigated only recently, primarily using morphological and serological approaches with few reports of molecular identification [14–16]. A serological study conducted using the Svanovir® A. marginale-Ab ELISA kit (SVANOVA Biotech AB, Uppsala, Sweden), revealed 28% of blood samples from lactating cows in northern Vietnam to be positive for specific antibody against A. marginale [15]. In other studies, Anaplasma spp. were suspected in animal samples from southern and central Vietnam [14,16]. There remains a need for accurate species identification for better molecular taxonomic understanding of the infections by Anaplasma spp. in Vietnam. The present study provides molecular characterization of Anaplasma spp. in blood samples and further identification of the Anaplasma species present in dairy and indigenous cattle and domestic dogs by using 16S rDNA analysis. The single 16S rDNA sequences have been successfully explored for taxonomic clarification of Anaplasma spp. and Ehrlichia spp. in many endemic regions/countries [2,6,8,12,13,17].
Over 2 years (May 2015 to August 2017), 226 blood samples were collected from cattle and domestic dogs in 2 provinces of the Red River Basin in the north of Vietnam. In the upland Bavi district in Ha Noi (21°5′0″N/105°23′0″E), 78 samples were obtained from hybrid dairy cattle used for milk production, 66 from indigenous cattle and 14 from domestic dogs kept by the farmers. In the lowland Kim Thanh district of Hai Duong Province (20°56′0″N/106°19′0″E), 57 blood samples were taken from indigenous cattle and 11 from dogs (Fig. 1A). The ethical approval was approved by the National Institute of Malariology, Parasitology and Entomology (NIMPE) on behalf of the Ministry of Health, Vietnam. Appropriate permission was obtained from the commune authorities and consent was obtained from cattle and dog owners. Blood samples from the animals were taken by experienced technicians/veterinarians.

Sample collection sites and a micrograph of a stained blood smear from cattle. (A) Map showing the locations of Ha Noi and Hai Duong Provinces in northern Vietnam (solid circle symbols). (B) Anaplasma marginale in erythrocytes (arrows) in a blood sample from cattle. Giemsa stain (×1,000).
About 2 ml of whole blood was collected from each animal into vials containing ethylenediaminetetraacetic acid (EDTA) anticoagulant (Sigma-Aldrich Co. LLC, Saint Louis, Missouri, USA), and kept on ice during transportation to the laboratory. Blood smears were stained using Giemsa (Sigma-Aldrich) and observed with light microscopy at 1,000× magnification (Fig. 1B).
Total genomic DNA was extracted from the middle phase of 400 μl of blood after centrifugation, from those individual hosts with Anaplasma spp. present in their blood smears. DNA extraction was done using the GeneJET™ Genomic DNA Purification Kit (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA). Amplification of a portion of the 16S rDNA gene was performed by nested PCR using first the primer pair (forward EHR1 5′ GAACGAACGCTGGCGGCAAGC 3′; and reverse EHR2 5′ AG-TAYCGRACCAGATAGCCGC 3′). Two microlitres of the first PCR product was used for the second, nested, PCR using primer pair EHR3 (5′ TGCATAGGAATCTACCTAGTAG 3′) and EHR4 (5′ CTAGGAATTCCGCTATCCTCT 3′) [12]. Each PCR was done in a reaction volume of 50 μl containing 25 μl DreamTaq PCR Master Mix from Thermo Fisher Scientific, 2 μl of each primer (10 pmol/μl), 2 μl DMSO, 16.0 μl pure water, and 3 μl template (-10 ng/μl). Amplification was carried out in a MJ thermal cycler PTC-100 (MJ Research, Watertown, Massachusetts, USA), with denaturation at 94°C for 5 min, followed by 35 cycles, each of 94°C/30 sec, 52°C/30 sec (annealing), 72°C for 2 min (extension), followed 10 min/72°C (final extension). The final PCR product was 510–520 bp in length, of which a 500–501 bp portion was used for alignment for taxonomic identification.
Our 10 16S rDNA sequences of Anaplasma spp. were supplemented with 15 Anaplasma sequences from GenBank. A sequence from Ehrlichia canis was used as an outgroup for phylogenetic analysis. All 26 sequences were aligned in GENED-OC 2.7 (Available from: http://iubio.bio.indiana.edu/soft/molbio/ibmpc/genedoc-readme.html). A phylogenetic tree was inferred using the maximum likelihood method in MEGA7 with the general time reversible model (GTR+G+I). Support for each node was assessed using 1,000 bootstrap resamplings [18].
Positive Giemsa-stained blood smears showed Anaplasma spp. present within bovine erythrocytes, with A. marginale typically located near the membrane (Fig. 1B). This species was identified on the basis of 16S rDNA analysis in 5 indigenous cattle (3 in Ha Noi; 2 in Hai Duong Province) and 2 dairy cattle (Ha Noi). Anaplasma platys was detected in 2 indigenous cattle (1 in each province) and 1 dog (in Ha Noi Province) (Table 1). Sequences of all 7 Vietnamese samples of A. marginale were identical and differed by 0.2–0.4% from the reference sequences used (strains from China and South Africa). Three other Vietnamese isolates identified as A. platys differed by less than 0.2% from the reference strains from Germany and the Philippines (KP006397). The inter-specific distance between A. marginale and other Anaplasma spp. was 4.6–5.0% (with A. bovis), 2.8–3.6% (with A. platys), and 1.0–3.6% (with A. phagocytophilum).
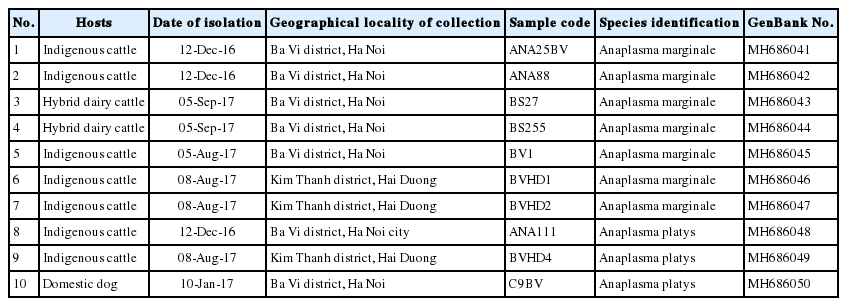
Molecular identity of Anaplasma spp. from indigenous and dairy cattle and domestic dog in 2 provinces in northern Vietnam
Phylogenetic analysis (Fig. 2) revealed the close relationship between the 7 A. marginale sequences from Vietnam and the reference strains from China, The Philippines, Zimbabwe and South Africa (99% bootstrap support). Similarly, the 3 Vietnamese A. platys sequences formed a well-supported group (90% bootstrap support) with 6 reference strains from China, Malaysia, Thailand, Germany, The Philippines and France. Anaplasma phagocytophilum (KT986058, South Korea; U02521, United States) and A. bovis (KY425445, Australia) formed separate branches in the tree, indicating a relatively distant relationship between A. marginale and A. platys (Fig. 2).

Phylogenetic tree showing taxonomic relationships of Anaplasma marginale detected from cattle and A. platys from cattle and dogs, in 2 provinces in northern Vietnam. The relationships were inferred based on 16S rDNA data (501–502 bp) from 26 sequences, including 10 from Vietnam and 15 reference sequences of Anaplasma species. Ehrlichia canis was used as an outgroup. Phylogenetic reconstruction was performed in MEGA7 using maximum likelihood analysis based on the general time-reversible model. Support for each node was indicated by 1,000 bootstrap resamplings [18]. The scale bar represents the number of substitutions per site. For each sequence, the full species names are followed up by strain abbreviations (in brackets) and the country (where they were isolated). The accession numbers are given at end of each sequence label. Ten Anaplasma spp. from Vietnam are marked with a solid circle. Names of hosts (cattle or dogs, in brackets) are added in the label for every sequence from Vietnam.
In this study, we have molecularly detected and identified Anaplasma spp., including A. marginale in the hybrid dairy cattle; A. platys in a domestic dog; and both species in the indigenous cattle. Phylogenetic tree showed the clear topology for 2 separate clusters of Anaplasma spp. of the Vietnamese samples indicating A. marginale and A. platys, respectively. Low genetic distance within strains in each species supported the taxonomic analysis.
A. marginale is specific to bovine hosts, infecting primarily lactating cattle, particularly hybrid cows, while A. platys has a wider host range that includes cattle, dogs and humans [3,15]. Our discovery of A. platys in cattle and domestic dogs raises considerable concern about zoonotic transmission in Vietnam, where indigenous cattle are grazed on grass fields and hybrid dairy cattle are kept in pens, fed daily by the farmers. Domestic dogs are commonly kept around farmers’ homes. This means very close contact between the host animals and the human population. It is important to plan future surveys, applying both morphological and molecular methods, for investigation of rickettsial bacteria in animals and humans in endemic areas through Vietnam.
In conclusion, our study has provided molecular species identification of Anaplasma marginale in hybrid dairy cattle and indigenous cattle, and of A. platys in cattle and dogs in 2 provinces in the Northern Vietnam. The presence of A. platys in cattle and dogs may constitute a potential zoonotic risk to humans. The systematic investigation of Anaplasma spp. and members of the Anaplasmataceae in animal hosts and humans is needed across Vietnam.
ACKNOWLEDGMENTS
This research was supported by the ARES-CCD funding (Académie de Recherche et d’Enseignement Supérieur). The authors thank the directorate and staff of Ba Vi Cattle and Forage Research Centre for sample collection. We express our thanks to colleagues and technicians in the Immunology Department, Institute of Biotechnology (IBT), Vietnam Academy of Science and Technology (VAST) for granting contribution to the molecular laboratory work. We extend our thanks to Prof. David Blair of James Cook University (Australia), for the invaluable review of the manuscript.
Notes
CONFLICT OF INTEREST
The authors declare no conflicts of interest in submission/publication of the data in this article.