Complete Sequence of the Mitochondrial Genome of Spirometra ranarum: Comparison with S. erinaceieuropaei and S. decipiens
Article information
Abstract
This study was undertaken to determine the complete mitochondrial DNA sequence and structure of the mitochondrial genome of Spirometra ranarum, and to compare it with those of S. erinaceieuropaei and S. decipiens. The aim of this study was to provide information of the species level taxonomy of Spirometra spp. using the mitochondrial genomes of 3 Spirometra tapeworms. The S. ranarum isolate originated from Myanmar. The mitochondrial genome sequence of S. ranarum was compared with that of S. erinaceieuropaei (GenBank no. KJ599680) and S. decipiens (Gen-Bank no. KJ599679). The complete mtDNA sequence of S. ranarum comprised 13,644 bp. The S. ranarum mt genome contained 36 genes comprising 12 protein-coding genes, 22 tRNAs and 2 rRNAs. The mt genome lacked the atp8 gene, as found for other cestodes. All genes in the S. ranarum mitochondrial genome are transcribed in the same direction and arranged in the same relative position with respect to gene loci as found for S. erinaceieuropaei and S. decipiens mt genomes. The overall nucleotide sequence divergence of 12 protein-coding genes between S. ranarum and S. decipiens differed by 1.5%, and 100% sequence similarity was found in the cox2 and nad6 genes, while the DNA sequence divergence of the cox1, nad1, and nad4 genes of S. ranarum and S. decipiens was 2.2%, 2.1%, and 2.6%, respectively.
Cestodes of the genus Spirometra (Mueller, 1934) are intestinal parasites of cats and dogs that use the fresh water copepod as the first intermediate host and reptiles or amphibians as the second intermediate host. Spirometra species have been reported from felids and canids worldwide under the generic name Diphyllobothrium since 19th century. Spirometra species were described from the morphological identification of 6 Spirometra species, comprising S. erinaceieuropaei (Rudolphi, 1819), S. decipiens (Diesing, 1850), S. ranarum (Gastaldi, 1854), S. mansoni (Cobbold, 1882), S. houghtoni (Syn. S. mansoni), and S. okumurai by Faust et al. [1]. Fifteen Spirometra species were reviewed as valid species and divided into 2 groups by Wardle and McLeod [2]. A Spirometra species in North America was reported as S. mansonoides (McIntosh, 1935) [3]. Five Spirometra species have been reported from wild felids in South America such as S. decipiens, S. mansoni, S. longicollis (Parodi and Widakowich, 1917), S. gracilis (Baer, 1927), and S. mansonoides [4]. S. pretoriensis and S. theileri (Baer, 1924) have been reported from a bush cat (Leptailurus serval) and a tiger cat (Felis lybica cafra) [5]. Currently, Spirometra species causing human sparganosis have been identified as S. erinaceieuropaei and S. decipiens by morphological and genetic analyses in Korea [6]. The most recent reports include identification of S. ranarum from frogs (Hoplobatrachus rugulosus) in Myanmar and from lions (Panthera leo) in Tanzania by analysis of mitochondrial genes and morphological observation [7,8]. S. ranarum obtained from cats and dogs were identified by molecular and phylogenetic analysis of DNA sequence data of the mitochondrial cytochrome c oxidase (cox1) gene in Korea [9].
A species level taxonomy of Spirometra species was employed based on morphological characteristics combined with molecular analysis. Mitochondrial DNA is considered to provide a useful molecular marker for taxonomic identification, inferences of phylogenetic relationships, population genetics and epidemiological investigation [10]. Mitochondrial DNA sequences have been studied for genetic variation, taxonomy, and phylogenetic relationships using cox1, cox3, nad1, nad3, and nad4 [10–14]. The complete mitochondrial genomes of order Diphyllobothridea have been published for D. latum, D. nihonkaiense, S. erinaceieuropaei, and S. decipiens [15–17]. This study was undertaken to determine the complete mitochondrial DNA sequence and structure of the mitochondrial genome of S. ranarum, and to compare it with those of S. erinaceieuropaei and S. decipiens. The aim of this study was to provide information of the species level taxonomy of Spirometra spp., by analysis of mitochondrial genomes of 3 Spirometra tapeworms.
The S. ranarum isolate originated from Myanmar. This Spirometra tapeworm was identified using morphological and molecular characteristics as well as animal inoculation experiments [7]. Specific identity was confirmed using sequences of the mitochondrial cox1 and nad1 genes previously reported for S. ranarum (GenBank No. MH298843, MH2998844). Mitochondria from a single specimen were isolated using a Qproteome Mitochondria Isolation Kit (Qiagen, Hilden, Germany). Mitochondrial DNA (mtDNA) was extracted from mitochondria using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, California, USA). Whole genome amplification (WGA) of extracted mtDNA was performed using an REPLI-g Mitochondrial DNA Kit (Qiagen, Hilden, Germany) combined with Exonuclease-Resistant Random Primer: 5′NpNpNpNpNpSNpSN-3′ (Thermo Scientific, Hudson, New Hampshire, USA). Amplified mtDNA was sequenced using an Illumina sequencing genome analyzer (Macrogen, Seoul, Korea). The mitogenome was reconstructed using the Geneious 9.0 software program (Biomatters Ltd, Auckland, New Zealand) and employing the partial cox1 gene of S. ranarum as initial bait. The complete mitochondrial genome was annotated and recovered using MITOS and Dogma [18,19]. The mitochondrial genome sequence of S. ranarum was compared with that of S. erinaceieuropaei (GenBank no. KJ599680) and S. decipiens (GenBank no. KJ599679). The 12 protein-encoding genes in the mtDNA was confirmed by comparison with mitochondrial gene sequences of other cestodes available in the GenBank database. Platyhelminth mitochondrial genetic code was used to translate the mitochondrial protein-coding genes. Twenty-two putative tRNA genes were identified using tRNAscan-SE.2.1 [20] and anticodon sequences. The putative stem-loop structures of non-coding regions were inferred using the RNAdraw program [21].
The complete mtDNA sequence of S. ranarum comprised 13,644 bp. The S. ranarum mt genome contained 36 genes comprising 12 protein-coding genes, 22 tRNAs and 2 rRNAs. The mt genome lacked the atp8 gene as with other cestodes. All genes in the S. ranarum mitochondrial genome are transcribed in the same direction and arranged in the same relative position with respect to gene loci as found for S. erinaceieuropaei and S. decipiens mt genomes (Table 1). The nucleotide composition of the entire mt genome of S. ranarum comprised 20.4% A, 11.2% C, 22.6% G, and 45.8% T (66.2% A+T). An A+T richness was observed in Spirometra species S. erinaceieuropaei (65.7% A+T) and S. decipiens (66.3% A+T) as with other cestodes. Some genes were found to overlap in the mitochondrial genome of S. ranarum: cox1/trnT (10 bp), nad4L/nad4 (39 bp), trnQ/trnF (3 bp), and trnF/trnM (3 bp) (Table 1).
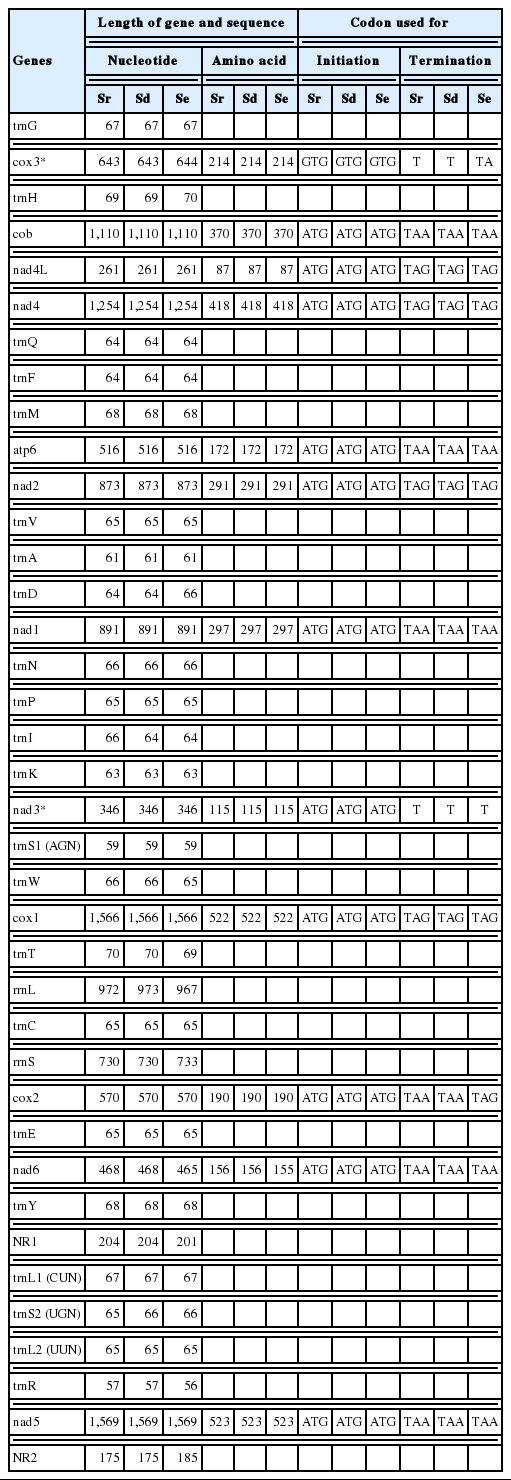
Position and characteristics of the protein-coding and non-coding sequences in the mt genome of Spirometra ranarum, S. decipiens and S. erinaceieuropaei
Accounting for some 74% of the entire mitochondrial genomes of Spirometra species are protein-coding genes. All putative open reading frames (ORFs) of the 12 protein-coding genes of S. ranarum mtDNA started and ended with complete codons. The ATG initiation codon was used in 11 genes (atp6, cob, cox1, cox2, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), while the GTG initiation codon was used only in the cox3 gene in S. ranarum, S. decipiens and S. erinaceieuropaei. The TAG stop codon was used in 4 genes (cox1, nad2, nad4, and nad4L), while the TAA stop codon was used in 6 genes (atp6, cob, cox2, nad1, nad5, and nad6) in S. ranarum and S. decipiens. The TAG codon was used in the cox2 gene in S. erinaceieuropaei. In cox3 and nad3, the abbreviated stop codon U was confirmed in S. ranarum and S. decipiens. The use of TA termination was found in cox3 for S. erinaceieuropaei mtDNAs. The most commonly used codons were for leucine, phenylalanine, valine and serine in mitochondrial proteins of 3 Spirometra species. The mitochondrial genome of Spirometra tapeworms appear to use the flatworm mitochondrial code, namely TTR and CTN for leucine, TTY for phenylalanine, GTN for valine, and AGN and TCN for serine.
Twenty-two tRNA genes were identified as putative secondary structures comprising a typical cloverleaf shape, with length ranging from 56 to 69 bp in S. ranarum and S. decipiens, respectively, and 56 to 70 bp in S. erinaceieuropaei. The inferred secondary structure of 19 tRNA exhibited a typical cloverleaf shape with 4 arms comprising aminoacyl acceptor arms, a DHU arm, anticodon stems and TΨC arms. trnR, trnS1, and trnS2 were replaced with 7–12 bp of unpaired loop in the DHU arms. The aminoacyl acceptor arms comprising 7 nt such as trnA, trnI, trnM, trnQ, trnR, trnS2, trnT, and trnV contained 1 or 3 non-canonical base pairs. The anticodon stems comprising 5 nt as with typical stem structures. The TΨC arms comprised a 2–5 nt stem with a 3–9 nt loop. The variable loop between the anticodon and the TΨC stems comprised 3–5 nt in 3 Spirometra species mtDNA. These 22 tRNAs had the same structure in the mitochondrial genome of other parasitic platyhelminths. Two mitochondrial ribosomal subunit genes rrnL and rrnS in the 3 Spirometra tapeworms were separated by trnC. The putative 16S rRNA and 12S rRNA genes in S. ranarum were 972 and 730 nt long, respectively. These sizes are similar to those of rRNA genes in S. decipiens and S. erinaceieuropaei, which range from 967 to 973 nt for 16S rRNA, and 730 to 733 nt for 12S rRNA (Table 1). The nucleotide content of the 16S rRNA and 12S rRNA genes in S. ranarum was 25.0% A, 12.5% C, 24.3% G and 38.1% T (63.1% A+T), whereas the A+T content was 63.1% in S. decipiens and 63.6% in S. erinaceieuropaei. Two major non-coding regions present in 3 Spirometra species mtDNA were predicted between trnY and trnL1, and between trnR and nad5. Non-coding region 1 (NR1) between trnY and trnL1, was 204 nt (S. ranarum), 204 nt (S. decipiens) and 201 nt (S. erinaceieuropaei) in length, while non-coding region 2 (NR2) between trnR and nad5 was 175 nt (S. ranarum), 175 nt (S. decipiens) and 185 nt (S. erinaceieuropaei) in length.
A percentage pairwise comparison of sequence divergence of the 12 protein-coding genes among S. ranarum, S. decipiens, and S. erinaceieuropaei is shown in Table 2. The 12 protein-coding genes constituted 10,067 bp and 3,355 codons (S. ranarum), 10,067 bp and 3,355 codons (S. decipiens), and 10,065 bp and 3,355 codons (S. erinaceieuropaei) of their respective mitochondrial genomes. The overall nucleotide sequence divergence of 12 protein-coding genes of S. ranarum and S. decipiens differed by 1.5%, while that of S. ranarum and S. erinaceieuropaei differed by 13.0%. The divergence of amino acid sequences of 12 protein-coding genes of S. ranarum and S. decipiens ranged from as low as 0.0% (cox2 and nad6) to as high as 1.4% (nad4) (Table 2). The nucleotide sequence divergence of the nad4 gene (the most variable gene), and the cox2 and nad6 (the most highly conserved gene) genes was 2.6% and 0.0% between S. ranarum and S. decipiens, respectively. The ribosomal RNA genes of S. ranarum and S. decipiens differed by 1.6% (16S rRNA) and 1% (12S rRNA). The percent of nucleotide sequence difference was greater than that of the amino acid difference for the cox1, cox3, cob, atp6, nad1, nad2, nad3, nad4, and nad4L genes of S. ranarum and S. decipiens, suggesting the existence of synonymous substitutions.
The degree of divergence in mtDNA sequences between sister species or congeneric species was estimated using the genetic distance of the cob gene among mammalian groups such as amphibians, reptiles and avian birds. The sequence divergence of the sister or congeneric species cob gene among mammalian groups is greater than 2% [22]. Another study reported that cox1 divergence among 13,320 species in 11 animal phyla ranged from as low as 0.0% to as high as 53.7%, while 79% of those species showed greater than 8% sequence divergence at the species taxonomy level [23]. The aforementioned studies have established that 98% of congeneric animal species showed greater than 2% sequence divergence in the cox1 gene [22,23].
Mitochondrial cox1 sequence variation of Spirometra species has been reported to range from 0.0–3.5% in China, Myanmar, Thailand, and Lao PDR, [24] and range from 0.0–2.6% in Japan, India and Indonesia [25]. In the present study, the complete mitochondrial genome of S. ranarum was sequenced and characterized, and comparison with that of S. erinaceieuropaei and S. decipiens showed that overall nucleotide sequence divergence of 12 protein-coding genes of S. ranarum and S. decipiens differed by 1.5%, with 100% sequence similarity in the cox2 and nad6 genes, while the DNA sequence divergence of cox1, nad1, and nad4 genes of S. ranarum and S. decipiens was 2.2%, 2.1% and 2.6%, respectively. These cox1, nad1, and nad4 genes have been used as genetic markers for taxonomic identification and phylogenetic reconstruction by many researchers [4,6–9,11–14,24,25]. The degree of sequence divergence of the cox1, cob, nad1, and nad4 genes of S. ranarum and S. decipiens was greater than 2%, which indicates that species are independent species. However, sequence differences of the cox3, atp6, nad2, nad3, nad4L, and nad5 genes of S. ranarum and S. decipiens was less than 2%, while that of the cox2 and nad6 genes was 0%, despite these 2 Spirometra species having morphological differences.
Although mitochondrial DNA sequence divergence has been useful as a genetic marker for species identification, including identification of sister or congeneric species, improving the reliability of this method requires the accumulation of DNA sequence data and criteria pertaining to morphological identification. A consensus has yet to be reached regarding criteria of mitochondrial DNA sequence divergence for species level taxonomy of Spirometra tapeworms, even though identified Spirometra species have been analyzed using mitochondrial DNA sequence divergence based on morphological characteristics. Further studies are needed to clarify species identification and to delineate the precise genetic variation of these Spirometra species.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (No. 2017R1D1A3B03035976). The materials used were provided by the Parasite Resource Bank of Korea (PRB000720).
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.
