Increased Innate Lymphoid Cell 3 and IL-17 Production in Mouse Lamina Propria Stimulated with Giardia lamblia
Article information
Abstract
Innate lymphoid cells (ILCs) are key players during an immune response at the mucosal surfaces, such as lung, skin, and gastrointestinal tract. Giardia lamblia is an extracellular protozoan pathogen that inhabits the human small intestine. In this study, ILCs prepared from the lamina propria of mouse small intestine were incubated with G. lamblia trophozoites. Transcriptional changes in G. lamblia-exposed ILCs resulted in identification of activation of several immune pathways. Secretion of interleukin (IL)-17A, IL-17F, IL-1β, and interferon-γ was increased, whereas levels of IL-13, IL-5, and IL-22, was maintained or reduced upon exposure to G. lamblia. Goup 3 ILC (ILC3) was found to be dominant amongst the ILCs, and increased significantly upon co-cultivation with G. lamblia trophozoites. Oral inoculation of G. lamblia trophozoites into mice resulted in their presence in the small intestine, of which, the highest number of parasites was detected at the 5 days-post infection. Increased ILC3 was observed amongst the ILC population at the 5 days-post infection. These findings indicate that ILC3 from the lamina propria secretes IL-17 in response to G. lamblia, leading to the intestinal pathology observed in giardiasis.
INTRODUCTION
Giardiasis is a gastrointestinal disease caused by Giardia lamblia, an extracellular protozoan [1]. This pathogen is ingested as an infective cyst, which is converted into trophozoites in the small intestine of the host. The resulting trophozoites are present either as free/swimming or attached to the surface of the epithelium, without invading the tissue. A distinct pathology of chronic giardiasis is shortening of the intestinal villi of the infected host [2]. Therefore, the mechanism by which these extracellular trophozoites trigger immune responses in the gastrointestinal tract of the host has been a subject of interest.
Diverse innate immune responses are activated in the small intestine infected with Giardia, which includes production of antimicrobial peptides [3], and nitric oxide (NO) [4] by intestinal epithelial cells. Interestingly, G. lamblia trophozoite has a strategy to evade the NO-mediated killing by actively utilizing arginine, a substrate for NO production [5]. Several cytokines had been reported to be up-regulated during Giardia infection, which include IL-6 [6], IL-17, IFN-γ, tumor necrosis factor-α and others. Especially, an in vitro stimulation of the host cells with Giardia extracts, induced release of IL-17 [7]. An in vivo infection model, using cattle, indicated a proliferation of IL-17-producing CD4+ T-cells [8]. Activation of IL-17-producing memory CD4+ T-cell was also demonstrated in G. lamblia infection in humans [9]. Role of IL-17 in Giardia clearance was proven using IL-17 receptor A (IL17RA) knockout (KO) mice [10]. Comparative microarray analysis of wildtype versus IL17RA KO mice provided information on IL-17 function in Giardia clearance, which included production of antimicrobial peptides and complement factors [11].
Gastrointestinal tract is the complex system which is constantly exposed to various exogenous organisms, diverse antigens, and ligands, not to mention with the normal microflora. Therefore, this system is required to maintain homeostasis and immune tolerance via complex and pleiotropic interactions among the epithelial and immune cells. Among them, the innate immune cells lacking specific antigenic receptors can be divided into myeloid cells and ILCs. In contrast to myeloid cells expressing pattern recognition receptors, ILCs are small molecule, like T-cell or B-cells, but lacking antigenic receptors. Therefore, ILCs response directly to cytokine from both myeloid cells and non-immune cells (such as epithelial cells), and transmit the information to other immune cells such as dendritic cells (DCs), T-cells or B-cells, eventually establishing both innate and adaptive responses in the hosts [12].
In this study, ILCs were isolated from the murine small intestine, and stimulated with Giardia trophozoites in culture plates containing transwell membranes. Comparative transcriptome analysis provided information on up-regulated genes of murine ILCs by G. lamblia, including diverse components of the immune response, one amongst which was the IL-17 pathway. By monitoring marker cytokines, the ILC group involved in G. lamblia-induced immune response, was determined via both in vitro and in vivo infection assays.
MATERIALS AND METHODS
Cultivation of G. lamblia
Trophozoites of G. lamblia GS strain (ATCC #50581, ATCC, Manassas, Virginia, USA) were grown under axenic conditions in a TYI-S-33 medium (2% casein digest, 1% yeast extract, 1% glucose, 0.2% NaCl, 0.2% L-cysteine, 0.02% ascorbic acid, 0.2% K2HPO4, 0.06% KH2PO4, 10% calf serum, and 0.5 mg/ml bovine bile, pH 7.1) [13] at 37°C.
Mice
C57BL/6 mice (6 week-old) were purchased from OrientBio Co. (Seongnam, Korea), and maintained at the animal facility of Yonsei University College of Medicine under SPF conditions. All experiments were approved by the Yonsei University College of Medicine Ethics Committee (IACUC 2017-0015).
Isolation of innate lymphoid cells
Mouse ILCs were prepared as described [14]. Briefly, the small intestines were extracted from C57BL/6 mice. After removal of the surrounding mesentery and Peyer’s patches, and fecal matters, the small intestines were minced into longitudinal pieces of 7 cm, and further cut into 1 cm pieces, followed by vigorous washes in phosphate buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4). They were further treated with 1 mM EDTA/PBS at 37°C for 20 min for removal of the intraepithelial lymphocyte and intestinal epithelial cell fractions. Thereafter, the samples were harvested by centrifuging for 20 min at 500×g, followed by vigorous washing for 20 sec in PBS. Further, they were minced into 1–2 mm pieces, and incubated in 10 ml of pre-warmed RPMI medium supplemented with 2% fetal bovine serum containing 20 μg/ml Liberase DH (Roche, Basel, Switzerland) and 50 μg/ml DNase I (Roche) at 37°C for 30 min. The cells were subsequently passed through a 70 μm EASY strainer (BD Biosciences, San Jose, California, USA), and a 37 μm nylon filter (BD Biosciences). The flow-through cells were harvested by centrifuging for 5 min at 500×g, 4°C, and then used as an ILC fraction.
ILC activation with G. lamblia tophozoites
ILCs (1×106 cells per well) were seeded in a 24 well culture plate with transwell membranes (Merck, USA) and incubated for 6 hr for RNA preparation and 16 hr for cytokine determination, with G. lamblia having multiplicity of infection (MOI) of 5. The culture media were collected for cytokine determination, and the ILCs were harvested for RNA preparation.
RNA isolation and expression analysis in RNA-sequence
RNAs were prepared from 4 independent ILC preparations, incubated with G. lamblia, and 4 control ILCs (ILCs only) using TRIzol (Invitrogen Life Technologies, Carlsbad, California, USA), and then treated with the RNase-free DNase (Qiagen, Valencia, California, USA) according to the manufacturer’s instructions. These RNA samples were submitted to Macrogen (Seoul, Korea) for RNA sequencing by HiSeq4000 (Illumina Inc., San Diego, California, USA). The resulting sequences were mapped against the genomic DNA reference (UCSC mm10) using HISAT2 program (Johns Hopkins University, USA). After alignment, the relative abundance of the transcripts was measured with the String Tie software (Johns Hopkins University), which measures abundance of each transcript as FPKM (Fragments Per Kilobase of transcript per Million mapped reads). The 4 samples for each group (ILCs alone or ILC incubated with G. lamblia) were analyzed as a cluster to monitor similarity within the group. The numbers of clusters were independently assessed using Hierachical clustering analysis (Euclidean method, Complete Linkage) to construct a heat map. Differentially expressed transcripts were annotated and automatically categorized using KEGG pathway analysis program (http://www.kegg.jp/kegg/pathway.html). The generated in the RNA-seq is available at the Gene Expression Omnibus (GEO) database with the accession number GSE130256.
Enzyme-linked immunoassay (ELISA)
Culture media from the ILCs incubated with G. lamblia, were used for detection of IL-17A, IL-17F, IL-22, IFN-γ, IL-1β, IL-5, and IL-13, by a sandwich ELISA to detecting IL-17A, as recommended by the manufacturer (BD Biosciences). The culture media collected from the ILCs, not exposed to G. lamblia, were used as controls for the assay.
Quantitative real-time PCR (qRT-PCR)
cDNA was synthesized from 4 μg of RNA using Prime-Script RT reagent kit (TaKaRa, Otsu, Japan) as directed by the manufacturer. The cDNAs were analyzed by qRT-PCR on a Light Cycler 480 II real-time PCR system (Roche Applied Science, Mannheim, Germany) using Light Cycler 480 DNA SYBR green I master kit (Roche Applied Science). The amount of each transcript was estimated using specific pair of primers; GAPDH, IL-17A, IL-17F, IL-22, IFN-γ, IL-1β, IL-5, and IL-13. The primers were made as suggested [15]. GAPDH gene was used as an internal control to normalize each transcript. Data analysis was based on the relative quantification method by determining the crossing point (Cp) value using the Light Cycler 480 II real-time PCR system software program (Roche Applied Science, version LSC480 1.5.0.39).
FACS
The collected ILCs were analyzed using the live/dead fixable dead cell stain kit (Invitrogen, Carlsbad, California, USA) to determine the percentage of dead (stained) cells. The process was followed by manufacturer’s instruction. Briefly, cells were pre-treated with anti-CD16/CD32 antibodies, and then cells were incubated with the following antibodies: FITC-conjugated anti-mouse Lineage negative (Lin-), AlexaFluor 700-conjugated anti-CD103, phycoerythrin (PE)-conjugated anti-IL-25R, allophycocyanin (APC)-conjugated anti-mouse I-A/I-E (MHC class II), AlexaFluor 700-conjugated anti-IL-17A or relevant isotype control antibodies in FACS buffer (PBS, 1% BSA, and 0.1% sodium azide). Fluorescence was measured by flow cytometry, and data were analyzed using FlowJo data analysis software (TreeStar, Ashland, Oregon, USA). All antibodies were from eBioscience (San Diego, California, USA) unless stated otherwise.
Mice infection with G. lamblia trophozoites
Each C57BL/6 mouse was infected orally with 5×105 G. lamblia trophozoites. Similarly, control mice were orally inoculated with 0.1 ml PBS, without G. lamblia trophozoites. The mice were sacrificed at various time-points, from 1 to 11 days post-infection (n=10), and their intestinal samples were collected for experimental purpose. Sections of duodenum paraffin blocks were stained with hematoxylin and eosin (H&E) and observed under Axiovert 200 fluorescence microscope (Carl Zeiss, Jena, Germany).
In addition, 10 mice were orally infected with G. lamblia as described, and their small intestines were removed at 5 days post-infection. The same number of control mice were necropsied and pooled as one group at the end of the trial. The small intestines obtained from mice of both groups were used to isolate ILCs as described above. These ILCs were analyzed for specific markers of the ILC group, using flow cytometry.
Statistical analysis
Results are expressed as means±standard deviations of 3 independent experiments. Data were analyzed by pair-wise comparison using Student’s t-test (SYSTAT program, SigmaPlot version 9; Systat Software Inc., Chicago, Illinois, USA). Differences were considered significant if the P-value was<0.05. Data with P-value<0.01 are indicated with 2 asterisks, and data with P-values between 0.01 and 0.05 are indicated by a single asterisk.
RESULTS
Up-regulation of IL-17 signal pathway genes in the ILCs by G. lamblia
The gene expression profiles were compared between ILCs stimulated with G. lamblia and control ILCs, using RNA-seq. For each group, 4 samples were analyzed, and their similarity within the group was monitored (Fig. 1A). Except one sample of ILC group stimulated with G. lamblia, the one-way Hierarchical Clustering analyses indicated valid tendency in the gene expression profile within the group. By applying strict selection of FPKM, i.e., P-values<0.05 and fold change >2 for all genes, 472 genes were found to be up-regulated and 498 genes were found to be down-regulated in the ILC group stimulated with G. lamblia (Fig. 1B). These genes were categorized 16 groups, according to their putative functions, which were presented as percentages of all the up- and down regulated genes (Fig. 1C). Substantial number of genes showing altered expression encoded hypothetical proteins (20–23%) and metabolic enzymes (9–13%) in both groups (up- and down-regulated clones). Eight to eleven percentage of up- and down-regulated genes were found to be involved in immune response, respectively. In addition, the expression of signaling genes (10–20%) was found altered in the ILC group stimulated with G. lamblia. Interestingly, in this group, more cell cycle-related genes (15%) was found to be down-regulated (15%) rather than up-regulated (3%).
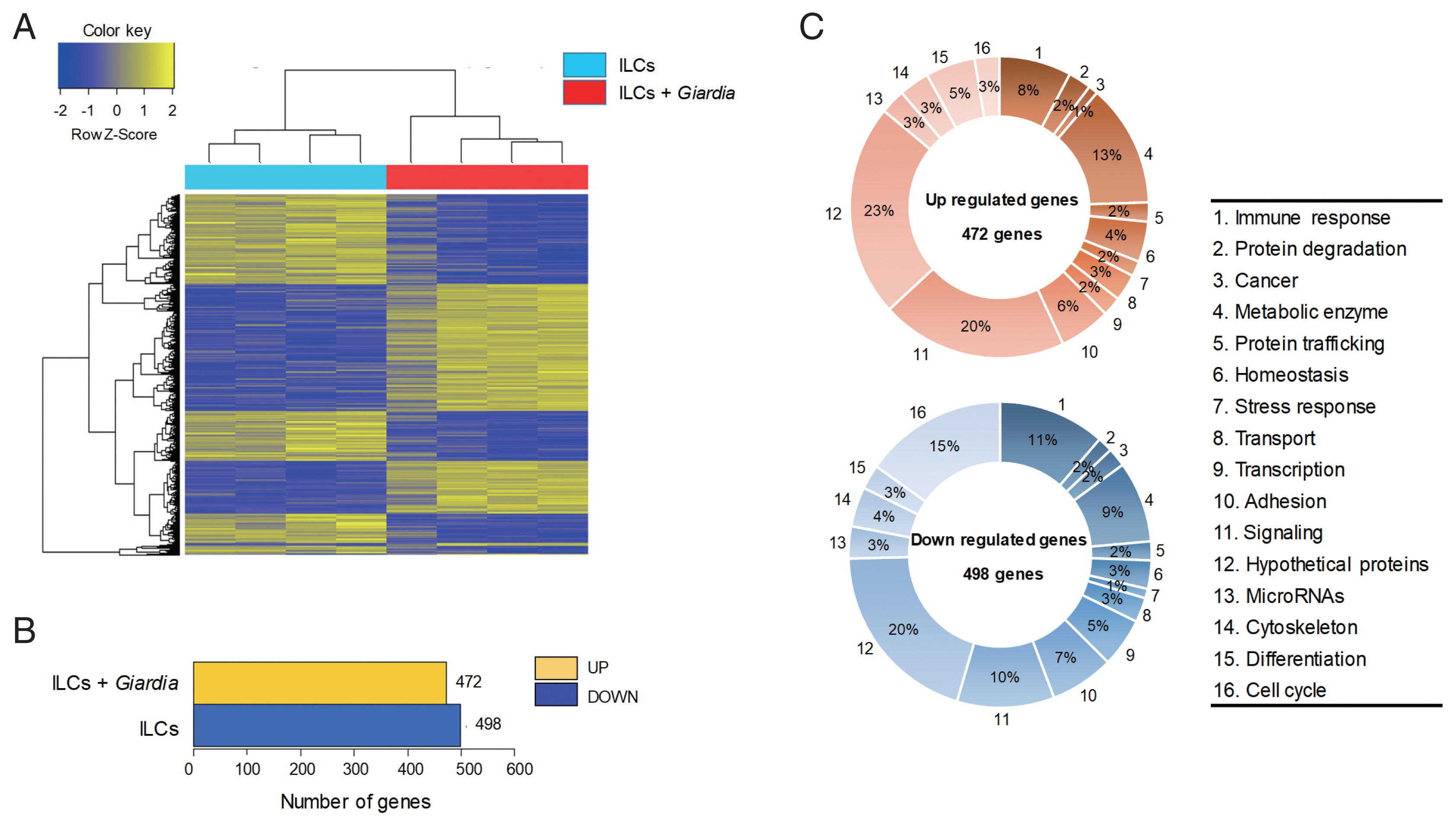
An RNA sequence analysis on the ILCs stimulated with G. lamblia. (A) Heat map across all the samples using the FPKM between the control ILCs and ILCs obtained after G. lamblia stimulation. Differentially expressed genes were sorted based on magnitude and sign of their t-statistics. (B) Number of up-, and down-regulated genes in ILCs incubated with G. lamblia with fold change more than 2 and P<0.05. (C) Diagrams showing the percentages of the up- or down-regulated genes in ILCs incubated with G. lamblia, categorized into their putative functions.
Especially, regulation of signature genes for diverse immune response was found among the modulated genes in these RNA-seq data. As expected, several signaling pathways seemed to be activated, which included chemokine signaling, MAPK signaling, Jak-STAT, PI3K-Akt, the Rap1 signaling pathways. In addition, the data suggested activation of several immune pathways, cytokine-cytokine receptor interaction, intestine immune response for IgA production, Th17 differentiation, Th1/Th2 differentiation, and IL-17 signaling pathway. Among the various pathways found to be activated by G. lamblia, we focused on the signaling pathway for IL-17, which has been earlier reported as a main regulator for immune response against G. lamblia [12].
Induction of IL-17 pathway related cytokines in vitro in the ILCs stimulated with G. lamblia
In this experiment, ILCs were incubated with G. lamblia in plates containing transwell membranes and examined for the secretion of the following cytokines: IFN-γ, IL-1β, IL-5, IL-13, IL-17A, IL-17F, and IL-22 (Fig. 2A). Secretion of IFN-γ, IL-1β, IL-17A, and IL-17F was found to be significantly increased. While there was moderate increase in IFN-γ, IL-1β, and IL-17F (2.5- to 3.5-fold), more than 15-fold increase in IL-17A secretion was observed. Secretion of IL-5, IL-13, and IL-22 was either decreased or not affected by G. lamblia.

Cytokine profiles of ILCs incubated with G. lamblia. (A) Cell-free supernatants were collected and assayed using ELISA kits. (B) Quantitative measurement of transcripts of cytokine genes. GAPDH was used as an internal control to normalize each transcript. *0.01<P<0.05, and **P<0.01 (Student’s t-test).
These data on cytokine secretion from ILCs stimulated with Giardia, were verified by qRT-PCR (Fig. 2B). Among the 7 cytokine genes, only the IL-17A and IL-17F transcripts showed a significant increase (2.4- and 2.7-fold). Moderate increase in the transcript of IFN-γ, IL-1β, and IL-22 was observed (1.4- to 1.7-fold). Decrease in the levels of IL-5 and IL-13 transcripts was noted.
Increase of ILC3 differentiation in vitro co-cultivation with G. lamblia
ILCs were divided into 3 groups: group 1 ILCs, group 2 ILCs, and group 3 ILCs, based on their cytokine production patterns that correspond to the helper T cell subsets, Th1, Th2, and Th17, respectively [12]. To determine which group of ILCs plays a role in G. lamblia-induced immune response, prepared murine ILCs were treated with Giardia, and assayed for expression of ILC markers using flow cytometry (Fig. 3A). In case of control cells, ILCs without G. lamblia stimulation, only a small portion of cells (0.02–0.3%) were stained with ILC1 (CD103) or ILC2 marker (IL-25R). The expression of these markers was increased to 0.2–2.3% when ILCs were stimulated with G. lamblia. Without G. lamblia stimulation, a significant portion of ILCs (25%) were stained with MHCII, an ILC3 marker. Expression of this surface protein was further increased to 53% when ILCs were incubated with G. lamblia. Along with IL-17 production, the FASC analysis indicated that among the 3 ILC groups, ILC3 played the most important role in G. lamblia-induced immune response.
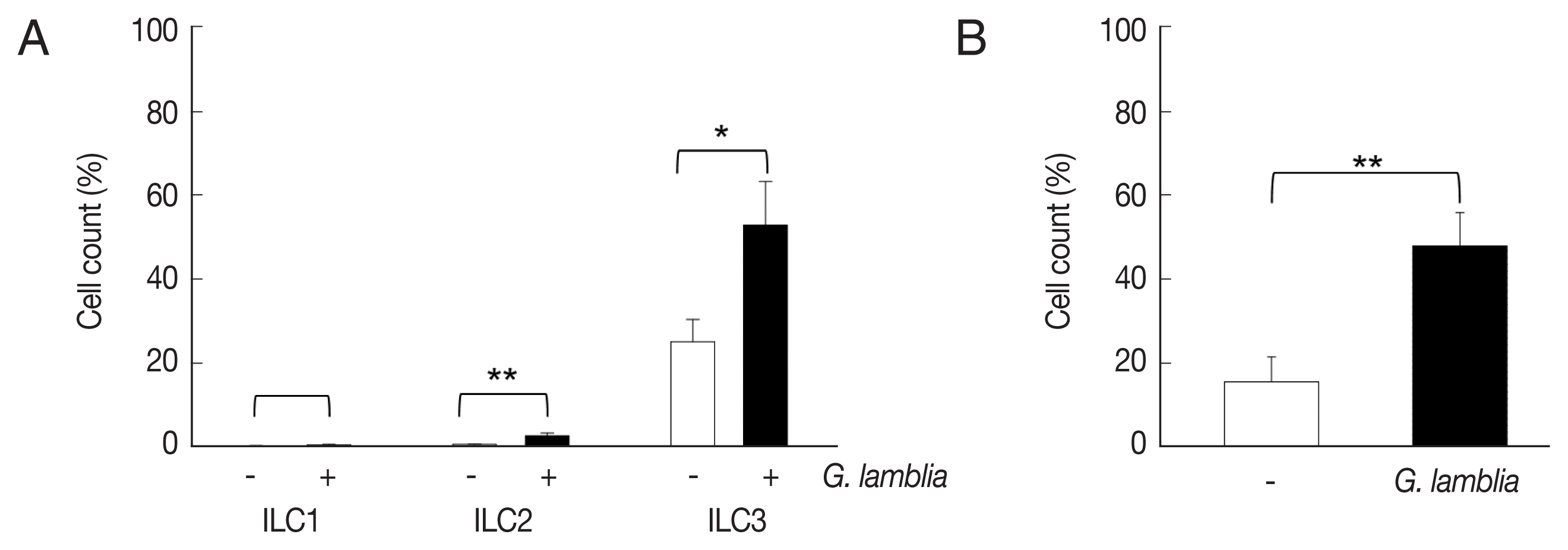
The activated group ILCs in the cells treated with G. lamblia in vitro. (A) Bar graph showing percentage of ILCs stained with markers for 3 group ILCs. (B) Bar graph showing percentage of cells expressing 2 ILC3 markers analyzed by FACS. *0.01<P<0.05, and **P<0.01 (Student’s t-test).
Therefore, increase of ILC3 in G. lamblia-treated cells was demonstrated by FACS analysis using MHCII and IL-17A, markers for ILC3 (Fig. 3B). Co-expression of these 2 proteins was observed in only 15% of the control cells (ILCs untreated with G. lamblia), while in 46% of ILCs stimulated with G. lamblia.
Increase of ILC3 differentiation in mice infectedwith G. lamblia
Role of ILC3 in G. lamblia-induced cytokine production was examined in vivo using a mouse model. Since mice are not natural hosts for G. lamblia, infection by G. lamblia trophozoites was monitored for 11 days. Small intestines of the infected mice were extracted and prepared for H&E staining for observation of G. lamblia trophozoites, using microscopy (Fig. 4A). On the first day, few trophozoites were found in the murine intestine. Maximal number of Giardia trophozoites was observed on the 5th day post-infection, and their presence gradually decreased, till no trophozoite was observed on days 10 and 11.
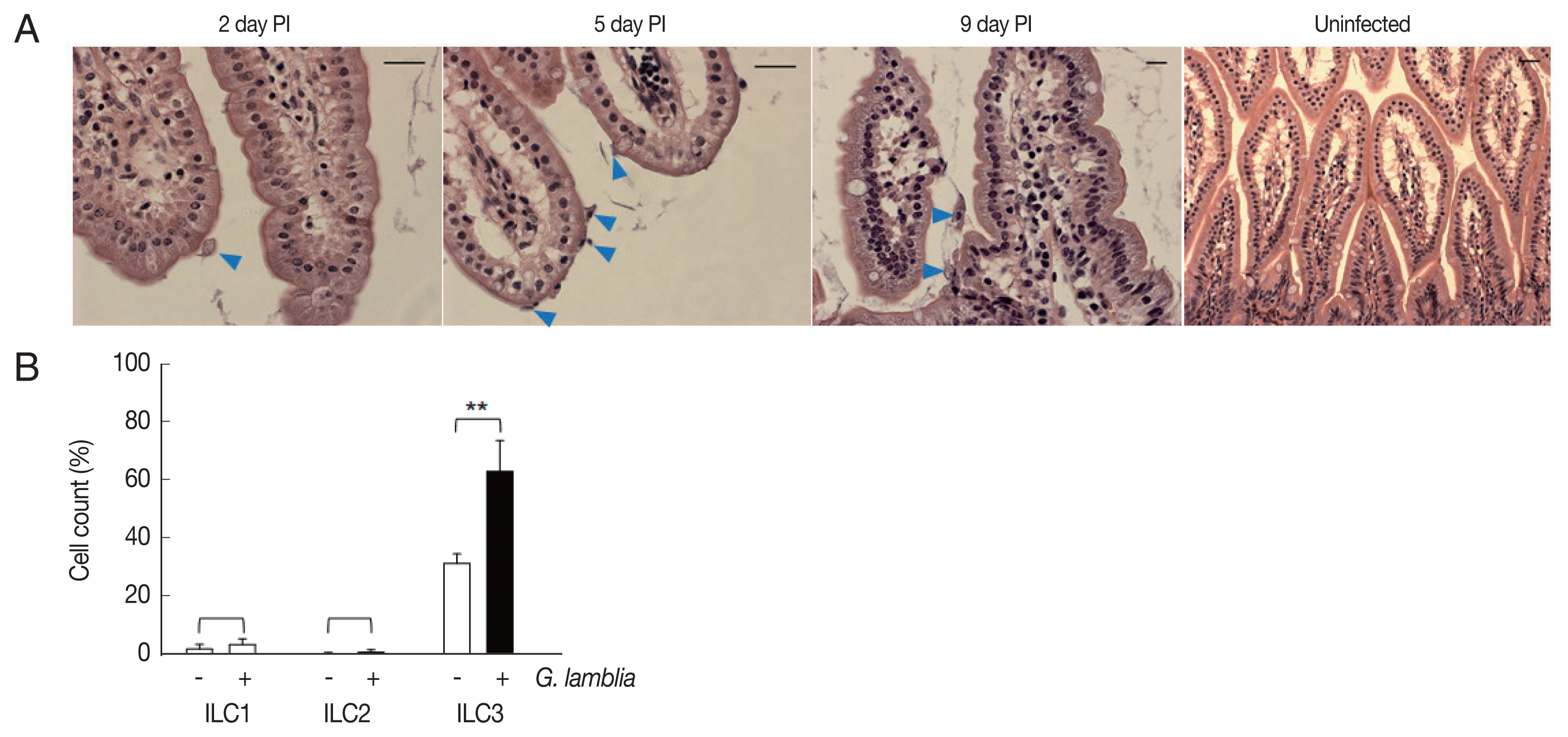
Activated group ILCs in the small intestine of mice infected with G. lamblia. (A) Small intestine of mice infected with G. lamblia. Arrowhead indicates G. lamblia trophozoites on the intestinal villi. PI; post-infection. Scale bar=50 μm. (B) Bar graph showing percentage of ILCs stained with markers for 3 group ILCs analyzed by FACS. *0.01<P<0.05, and **P<0.01 (Student’s t-test).
Based on the infection kinetics of mice with G. lamblia, ILCs were prepared from the mice infected with G. lamblia, at 5 days post-infection, and the 3 groups of ILCs were studied using flow cytometry (Fig. 4B). Less than 1% of the cells (0.11%) were stained, either with ILC1 or ILC2 marker, in the control ILCs, isolated from the uninfected mice. ILCs obtained from G. lamblia-infected mice showed 1.9% and 0.2% staining for ILC1 and ILC2, respectively. Corroborating with our in vitro data, more ILCs were stained with the ILC3 marker, both from the uninfected mice (35%) as well as from the G. lamblia-infected mice. However, the percentage of the ILC3-positive cells increased to 66% in the ILCs isolated from mice infected with G. lamblia, indicating a role of ILC3 in G. lamblia-induced immune response, which occurred in intestine of the infected mice.
DISCUSSION
The gastrointestinal tract is the primary site where G. lamblia initiates its infection. Among the various cells present in the small intestine, the ILCs were specifically studied to investigate how they modulate the immune response and adapt to G. lamblia trophozoites. Murine ILCs were prepared and challenged with G. lamblia trophozoites in vitro, resulting in death of ILCs (data not shown). Thus, ILCs were incubated with Giardia trophozoites in the presence of a membrane, such that only excretory-secretory products (ESPs) of Giardia could reach the ILCs, without inducing any cytotoxicity against them.
Transcriptional changes were monitored at the genomic level by comparative RNA-seq analysis of ILCs versus ILCs treated with Giardia ESPs (Fig. 1). For each group, 4 independent RNA samples were analyzed. All of 4 control samples (ILCs) demonstrated a meaningful consistency in the expression level of the whole genomes, whereas only 3 RNAs derived from ILCs with Giardia exposure showed similar pattern of gene expression. When both up-regulated and down-regulated genes were categorized by their putative function (Fig. 1C), significant numbers of regulated genes (8–11%, and 10–20%) were found to encode immune regulators, and components of signaling pathways, respectively, as expected from function of the ILCs. Based on transcriptional changes of the cytokines, chemokines, and their cognate receptors in ILCs [16], we could speculate that Th1, but not Th2, differentiation occurred in ILCs exposed to Giardia. However, more elaborate RNA-seq data should be obtained from ILCs incubated with Giardia for various time-points in order to draw a conclusion in this hypothesis.
Interestingly, the RNA-seq data revealed that the expression of numerous cell-cycle related genes (15% of the down-regulated genes) were decreased in ILCs incubated with Giardia. This result indicates that even though Giardia ESPs did not induce death of ILCs, they certainly altered the normal process of cell cycle of the ILCs.
Among candidate pathways activated in ILCs stimulated with Giardia, we noticed the IL-17 signaling pathway, of which thirteen signature components are found to be regulated. IL-17 is a well-known marker cytokine for group 3 ILCs [17], and has been reported as a pro-inflammatory cytokine involved in Giardia-induced immune responses [18]. Marker cytokines for ILC1 and ILC3 were detected by ELISA in the supernatants released from ILCs with Giardia (Fig. 2A). Among the 3 ILC3 cytokines, IL-22 was not detected in ILCs stimulated with Giardia, contradicting a previous study, reporting that most of the ILC3 cells (both Np46+ and Np46−) produced IL-22 [19]. However, increased il-22 gene transcripts were detected by qRT-PCR (Fig. 2B). In this assay, all the ILC1 and ILC3 marker genes showed elevated transcript levels in the ILCs challenged with G. lamblia. On the contrary, 2 ILC2 markers, IL-5 and IL-13, did not show any up-regulation in this group via both ELISA as well as qRT-PCR. These data suggest that ILC2 is not the main group ILC responsible for Giardia-induced immune response.
An assay to monitor relative expression of ILC surface markers (Fig. 3) clearly demonstrated that the ILC fraction prepared in this study had more ILC3 than ILC1 and ILC2. Abundance of ILC3 in the gastrointestinal tract has been reported in mice [20]. On the contrary, ILC2 are normally known to reside in the lungs of naïve animals [21]. Even though all 3 ILCs were found to be increased upon exposure to G. lamblia, significant increase of the ILC3 (46–53%) population was observed in the ILCs treated with Giardia, as shown in experiments using single- and double-staining of ILC3 markers. Based on previous studies that have shown that ILC3 can modulate CD4+ T-cell responses through antigen (Ag)-peptide presentation by MHC class II [12,22], it is hypothesized that ILC3- and MHCII-dependent mechanism is also involved in the adaptive host immune responses to G. lamblia.
To examine whether ILC3 activation observed in the in vitro assays using ILCs, actually occurs in vivo during G. lamblia infection, G. lamblia trophozoites were orally inoculated into mice, and the infection was monitored (Fig. 4A). Since mice are not natural hosts for G. lamblia, the infection did not result in shedding of Giardia cysts, and therefore, was monitored by observing the presence of Giardia trophozoites in the small intestine. As reported by other groups [10], the highest number of trophozoites was observed at day 5, and decreased thereafter. ILCs prepared from infected intestine at day 5 showed increased expression of ILC3 marker, corroborating the findings of our in vitro experiments (Fig. 4B).
These results suggest a role of ILC3 in G. lamblia-induced immune response in the small intestine of the host. Previous reports have indicated an important function for ILC3 in regulating interaction between the gastrointestinal tract and pathogens such as Citrobacter rodentium [23]. On the contrary, ILC2 was found to be responsible for the immune response against helminths and lung diseases, such as asthma [24].
The function of ILC3 in G. lamblia-triggered immune response in the gastrointestinal tract is complex and challenging to decipher, requiring elaborate investigations on the function of multiple immune and intestinal epithelial cells, as well as crosstalks among them. For example, in mice having ILC3 with impaired MHC class II, both Ag-specific T-cell and T-dependent B-cell responses were affected, showing that ILC3 plays a role in Ag presentation to CD4+ T-cells in vivo [25]. In addition, lymphotoxin-activated ILC3 was shown to modulate T-cell independent IgA induction via NO production by CD11c+ DCs [26]. Therefore, ILCs may have a critical role not only in modulating cytokine-mediated epithelial cell barrier integrity, but also in regulating adaptive immune cells. Future studies should focus on downstream cells of ILC3 and their effect on the immune response against G. lamblia.
ACKNOWLEDGMENT
This work was supported by the Research Fellow Program through National Research Foundation of Korea (NRF) grant funded by the Korea government (No. 2016R1A6A3A11934451).
Notes
CONFLICT OF INTEREST
The authors have no financial conflicts of interest to declare.