Abstract
We tried a series of morphological and molecular approaches to identify a new species of Stellantchasmus (Digenea: Heterophyidae) originating from the wrestling half-beaked fish, Dermogenys pusillus of Thailand. Adult worm samples of the new species were recovered from hamsters experimentally infected with the metacercariae from D. pusillus in Thailand. Two isolates (Thai and Korean) of Stellantchasmus falcatus were used as comparative control groups. Worm samples of 3 Stellantchasmus groups were morphologically observed and molecularly analyzed with the mitochondrial cytochrome c oxidase 1 gene. The morphological characteristics of S. dermogenysi n. sp. are similar to S. falcatus originating from brackish water fish, but minor difference was noted including the absence of the prepharynx, position of the ovary near the ceca end, smaller body size, and shorter esophageal length. A phylogenetic tree derived from neighbor-joining and maximum-likelihood methods suggests that S. dermogenysi n. sp. is separated from S. falcatus supported by high bootstrap values. The relative divergences persist between these host-specific trematodes, which we suggest should be recognized as 2 distinct species. Comparisons of S. dermogenysi n. sp. with S. falcatus isolated from mullets in Thailand and Korea indicate a genetic divergence of mitochondrial DNA of 19.4% and 21.7%, respectively. By the present study, a new species, Stellantchasmus dermogenysi n. sp. (Digenea: Heterophyidae), is proposed in Thailand based on molecular evidences, in addition to minor morphological differences between S. falcatus and the new species.
-
Key words: Stellantchasmus dermogenysi, Stellantchasmus falcatus, heterophyid fluke, phylogeny, Thailand
INTRODUCTION
Stellantchasmus is a genus of minute intestinal flukes from the family Heterophyidae. This genus includes only 4 known species:
S. falcatus Onji and Nishio, 1924,
S. aspinosus Pearson, 1964,
S. gallinae Oshmarin, 1970, and
S. batillans Pearson, 1964, all of which have been clearly defined by their morphological characters [
1]. The number and arrangement of testes and the presence of an armed or unarmed ventral sucker are used as the main characters for species identification [
1].
Stellantchasmus has been found in both experimental and natural hosts:
S. falcatus in natural and experimental cats;
S. aspinosus in the wild water rat and the experimental cat, rat, and chicken;
S. gallinae in the domestic chicken; and
S. batillans in the water rat and domestic duck [
1,
2].
S. falcatus was first reported in 1916 by Onji and Nishio [
3]. Human infection by this parasite has been reported in Japan, the Philippines, the Republic of Korea (=Korea), Hawaii, and Thailand [
4–
7], and human infections also occur at a high rate in northern parts of Thailand [
8]. So far only 1 species,
S. falcatus, has been recorded in Thailand (and in some other countries) [
4–
8]. During infection, eggs can pass through the blood vessels to the brain or heart of the patient [
9]. Pathology in humans includes mild to colicky pain, leading to death in severe cases (as seen in the Philippines) [
7,
9].
S. falcatus is widespread and can infect various types of hosts [
3,
10]. The first intermediate host of this parasite, reported in Thailand, is freshwater snails such as
Thiara scabra and
Tarebia granifera [
11]. The metacercarial stage of
S. falcatus, which infects humans, is found in several types of brackish water and freshwater fish, the second intermediate host [
10,
12]. There are only 2 species of fish,
Dermogenys pusillus (half-beaked fish, freshwater species) and
Planiliza subviridis (a species of mullet, brackish water species), that were reported to be second intermediate hosts of
S. falcatus in Thailand [
12]. Traditional identification of
Stellantchasmus species has been based solely on morphological characteristics; however, numerous morphological similarities can result in misidentification.
Over the last decade, cryptic speciation has been discovered in other digenean parasitic species such as
Echinostoma, Phyllodistomum, and
Clinostomum [
13–
17]. Mechanisms of speciation in parasites may be unswervingly connected to their hosts and their complex life cycles [
13,
18]. The presence of cryptic species arises through morphological similarities and appears to be widespread among parasites [
18,
19]. Speciation almost always occurs in parasite species that form a large diverse group of organisms, infecting various kinds of hosts [
13,
17,
20]. There have been reports of morphological characteristics of
S. falcatus from the fish
D. pusillus that seems to be different from
S. falcatus from the fish
P. subviridis [
7,
9,
10,
12,
21]. In this case, no exception exists in the ordinary cryptic speciation that may occur in
Stellantchasmus species.
Molecular approaches are the most efficient tools currently used to understand phylogenetic relationships and to determine genetic variations in heterophyid flukes [
22–
25]. The conventional PCR and high annealing temperature-based random amplified polymorphic DNA (HAT-RAPD) methods have been used to discriminate
Stellantchasmus species isolated from various hosts [
22]. It has been suggested that different DNA fingerprints of
Stellantchasmus worms acquired from different hosts may be due to distinct species [
22]. To clarify this point, HAT-RAPD can be used to develop specific markers to detect
S. falcatus infections [
25]. Moreover, many genes, such as nuclear genes (including internal transcribed spacers 1 and 2; ITS1 and ITS2) and mitochondrial genes (including mitochondrial cytochrome
c oxidase 1; CO1), have been used for identification and phylogenic studies of various parasites [
13,
23,
24]. ITS2 and CO1 regions have been useful for investigation and identification of
Stellantchasmus sp. [
23,
24].
The aim of this study was to investigate and identify a new species of Stellantchasmus (Digenea: Heterophyidae) obtained from the wrestling half-beaked fish, D. pusillus, in Thailand based on both morphological and molecular approaches using CO1 gene.
MATERIALS AND METHODS
Ethical statements
All experimental hosts were managed according to the guidelines approved by the Animal Ethics Committee of the Faculty of Science, Chiang Mai University, Thailand, and the relevant document (no. RE 001/13) was approved by the committee. The guidelines for animal care were used according to the International Guiding Principles of Biomedical Research Involving Animals of Council for International Organizations of Medical Sciences (CIOMS).
Morphological investigation
The metacercariae of
Stellantchasmus spp. were collected from different sites in Thailand. The hosts of this parasite were
P. subviridis and
D. pusillus, acquired from Chiang Mai and Chonburi Provinces in Thailand. Fish specimens were minced and digested using 1% pepsin solution for 2 hr at 37°C and rinsed with 0.85% NaCl. A hundred metacercariae of
Stellantchasmus spp. were checked with light microscopy, and hamsters (
Phodopus campbelli) were force-fed with the metacercariae samples. After 2 days, the adult-stage worms were collected from the hamsters. Total 57 specimens of
Stellantchasmus were used for identification and morphological studies. The worms were processed for permanent slides. Briefly, they were flattened under a cover slip pressure and fixed in 4% formalin, stained with hematoxylin and/or Borax’s carmine, dehydrated in alcohol series, and finally mounted in Permount. The specimens were checked with the descriptions of Onji and Nishio [
3], Pearson [
2], Kliks and Tantachamrun [
5], Pearson and Ow-Yang [
1], and Pubua and Wongsawad [
12]. The body and organs of the worms were measured and analyzed. The holotype and paratypes of the new species are kept in the Natural History Museum, London (NHMUK). The worms were frozen at −20°C for DNA extraction.
Genomic DNA from each Stellantchasmus species and related groups were collected from adult worms. S. falcatus from the laboratory of Prof. Jong-Yil Chai, Seoul National University College of Medicine and Korean Association of Health Promotion, Seoul, Korea, was used for molecular studies with the 2 Thai samples. Approximately, 2 mg of each trematode tissue sample was used for DNA extraction. The DNA extraction and purification steps were performed using 150 ml of 5% Chelex (Fluka) solution containing 10 μl of 20 mg/ml proteinase K (Sigma-Aldrich, St. Louis, Missouri, USA): the mixture containing the trematode tissue was washed and incubated at 55°C for 1 hr, followed by heating at 95°C for 30 min, then gently mixed. The mixture was centrifuged for 10 sec at 13,000 rpm, after which the supernatant was removed and stored at −20°C until use.
Amplification and sequencing of the CO1 gene
The partial CO1 gene was amplified using the primers as follows: JB3 (5′-TTTTTTGGGCATCCTGACGTTTAT-3′) as a forward primer and JB4.5 (5′–TAAAGAAAGAACATAATGAAAATG-3′) as a reverse primer. The PCR amplifications were carried out as described above. The amplification procedure included a 3 min initial denaturation at 95°C, then 40 cycles of 1 min denaturation at 95°C, 1 min annealing at 50°C, 1 min extension at 72°C, and a 7 min final extension at 72°C. The PCR products were visualized on 1.0% agarose gels, purified using the Cleanup PCR Kit (Sigma-Aldrich), and directly sequenced in both directions by Ward Medic, Ltd. (Bangkok, Thailand).
Phylogenetic analyses
The molecular data in this study including the sequences from
Stellantchasmus spp. and outgroup (
Metagonimus spp.) were aligned using ClustalW, a subprogram of MEGA version 6.0 [
26]. The sequences were checked for ambiguous nucleotide sites before being subjected to a phylogenetic analysis. The jModel test 2 [
27] was used to find the most appropriate substitution model for the
CO1 datasets. Phylogenetic trees of all taxa were constructed using neighbor-joining, maximum likelihood, and Bayesian inference. GTR+G model was applied for all analyses. The NJ analysis and the likelihood scores of dataset was carried out using PAUP* version 4.0b10 [
28] with 1,000 bootstrap replicates. The ML analysis was undertaken using PHYML version 2.4.5 [
29]. The tree searching system used for this method involved the heuristic procedure with tree-bisection-reconnection branch swapping. The bootstrap resampling [
30,
31] was done with 1,000 replicates that were assigned to support the particular branches. BI analysis was performed using MrBayes version 3.2.5 [
32], where the Bayesian analysis was run for 5 million generations (heating parameter=0.15), sampling was done for every 100 generations, and the first 25% of trees were discarded with burn-in procedure. The final consensus tree was built using the last 15,002 trees. Support for nodes was defined as posterior probabilities (P).
RESULTS
Morphological data
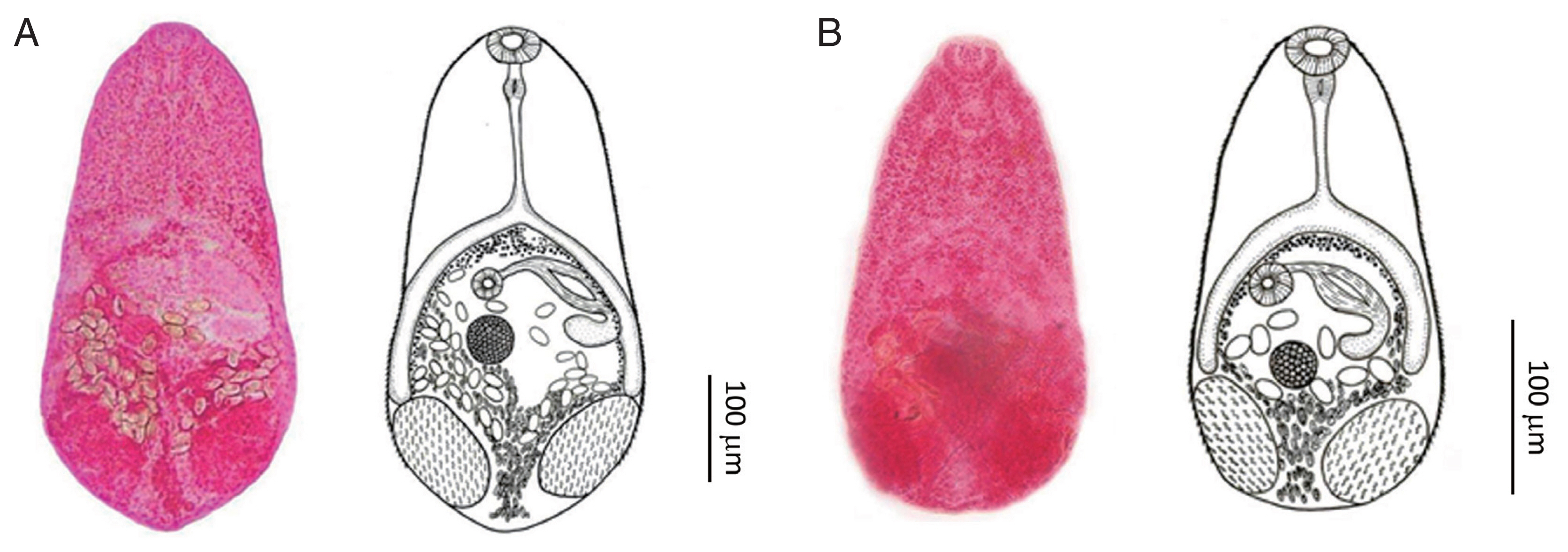
The samples of
S. dermogenysi were identified compared with the original description and holotype picture of
S. falcatus. In the morphology of
Stellantchasmus spp. from different hosts [
2–
4], the differences in the presence or absence of the prepharynx, position of the ovary, body size, and esophagus length were distinguishing factors. The prepharynx was very short or absent in
S. dermogenysi n. sp. but it was present in
S. falcatus. The ovary of
S. dermogenysi n. sp. is located near the ceca end, whereas in
S. falcatus it is located in the middle of the ceca length. Of the new species, the body, esophagus, and ventral sucker were clearly smaller than those of
S. falcatus (
Table 2;
Fig. 1). Similarity in the body shape, organ size, and their positions were observed, in addition to the ratios of the body size (
Table 2;
Fig. 1).
S. falcatus from
P. subviridis revealed smaller values. Although the ratio of the body size of
S. falcatus from
P. subviridis to the new species was 1.33:1.39, ratios of the organs were found to be different, especially with regard to the esophagus length, ovary width, and the right testis width (
Table 2).
Descriptions
Adult specimens were obtained from experimentally infected hamsters (P. campbelli). All measurements below are given in micrometers (μm). Body pyriform, with a length of 374 (256–480), and forebody narrower than the hindbody, with a maximum width of 167 (136–220) across the testes. Almost the entire surface covered with scale-like spines extending to half of the posterior body. Oral sucker subterminal and 36 (26–46) long by 42 (32–54) wide; prepharynx 4 (2–10) long; pharynx 27 (22–40) long by 25 (20–34) wide; esophagus 56 (30–102) long, with bifurcation extending to the anterior border of testis. Ventral sucker 22 (20–30) long by 24 (20–30) wide. Testes paired, ovoid, opposite, and in the posterior part of the body, and measured 90 (68–116) long by 58 (40–80) wide (right). Left testis 90 (68–112) long by 57 (40–82) wide. Seminal vesicles thin-walled, located to the left of ovary, and connected to the expulsor lying dorsal and left of cecum. Expulsor with muscle fibers 54 (26–80) long by 26 (20–36) wide, opened into the ventrogenital sac dorsally on the left side and anterior to ovary. Ovary slightly ovoid and positioned slightly right and anterior to testes, 53 (30–70) long by 49 (40–76). Uterus coiled in the posterior of the body and filled with operculate eggs. Vitelline follicles distributed at the inner intestine, with bifurcation extending to the posterior of the body. Eggs 21.6 (17.5–23.0) long by 11.5 (7.5–15.0) wide, thin-walled, possessing a smooth shell with a small operculum at anterior end.
Type materials
Type specimens have been deposited in the Natural History Museum, London, UK and the Applied Parasitology Research Laboratory, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand. Holotype NHMUK 2015.1.7.1 (
Fig. 1), paratypes NHMUK 2015.1.7.2 (1 individual) and CMUZ 01.1–01.28 (29 individuals).
Host
The metacercariae were collected from the freshwater fish Dermogenys pusillus and the adult stage in the experimental hamster (Phodopus campbelli).
Type locality
Chiang Mai Province, Thailand
Etymology
The name is derived from their specific second intermediate host, the wrestling half-beaked fish (Dermogenys pusillus).
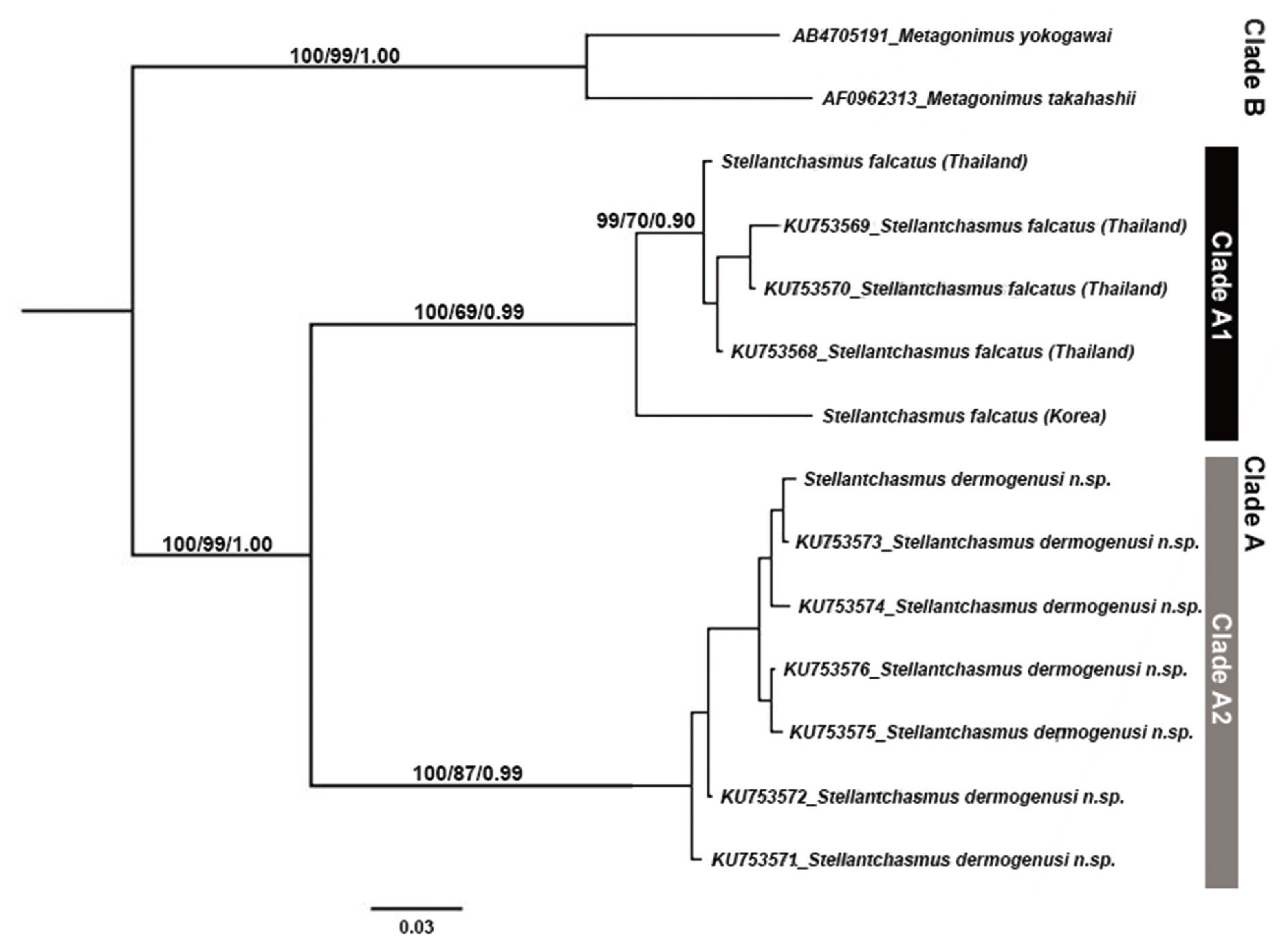
Phylogenetic analyses based on CO1 gene and genetic distance
The aligned 396 bp sequences of all the CO1 fragment codons had nucleotide frequencies of 0.173, 0.145, 0.290, and 0.392 for A, C, G, and T, respectively (41.1% GC content), with a 105 parsimony informative result and 126 variable sites. The K2P-distance between the taxa ranged from 0.005 to 0.217. Stop codons of all CO1 sequences were absent.
The phylogenetic trees were reconstructed using NJ, ML and BI methods (
Fig. 2). All the trees showed similar tree topology with high statistical support. The tree could be divided into 2 main clades. The clade A was the
Stellantchasmus group, and the clade B was the
Metagonimus group (outgroup). The clade A was also subdivided into 2 subclades, including
S. falcatus (clade A1) and
S. dermogenysi n. sp. (clade A2).
S. dermogenysi n. sp. (clade A2) was separated from both Thai and Korean
S. falcatus isolates which had high bootstraps supported (100% and 99% for NJ and ML bootstrap replicates and 1.00 for BI posterior probability, respectively). The distance among the
Stellantchasmus species complex is shown in
Table 3. The distance between
S. dermogenysi n. sp. and both Thai and Korean
S. falcatus was 0.194 (19.4%) and 0.217 (21.7%), respectively, while that between Thai and Korean
S. falcatus was lower (0.083 or 8.3%).
DISCUSSION
The occurrence of
S. dermogenysi n. sp. has been reported previously in northern Thailand, under the name
S. falcatus from the specific fish host
D. pusillus [
5,
7,
10,
12,
21]. Its morphology is clearly different from
S. batillans, which has a single testis. The arrangement of the testes is opposite in the new species, while in
S. gallinae it is in tandem position.
S. aspinosus, with an unarmed ventral sucker, is distinct from the new species, which has a ventral sucker with groups of tiny spines on the lip [
1]. The morphological measurements are similar to those reported for
S. falcatus from brackish water fish, but the difference in the length of the prepharynx, position of the ovary, body size, esophagus length, and different second intermediate host were distinguishing factors. The prepharynx is almost absent in
S. dermogenysi n. sp., but present in
S. falcatus, as described by Onji and Nishio in 1916 [
3], Pearson in 1964 [
2], and Chai et al. in 2016 [
33] and 2017 [
34], and in
S. falcatus from humans in Korea (used as comparative specimens in this study). In
S. dermogenysi n. sp., the ovary is located near the ceca end, whereas in
S. falcatus it is located mid-way along the ceca. The body size, esophagus length, and ventral sucker size of the new species are clearly smaller than in
S. falcatus. Most species of mullet fish serve as the second intermediate host for
S. falcatus, including
Mugil sp. in Korea [
10],
Chelon macrolepis in Cambodia [
33] and Myanmar [
34], and
P. subviridis in Thailand (from this study).
Stellantchasmus found in
D. pusillus, from Thailand, was also reported under the name
S. falcatus, but it was morphologically different from the
S. falcatus originating from mullets [
8,
22]. Their prevalence (100%) and intensity (22–2,492 metacercaria/fish) were very high in
D. pusillus [
8].
We used molecular methods to confirm
S. dermogenysi as a new species, and derived the name from their specific second intermediate host, the wrestling half-beaked fish (
D. pusillus). Mitochondrial genes, especially the
CO1 gene, have been widely used for identification of parasites and studies of phylogenetic relationships and genetic variations in the family Heterophyidae [
23]. In our study, the
CO1 gene revealed that the monophyletic tree of the
Stellantchasmus group (clade A) was clearly distinct from the related outgroup (
Metagonimus spp.), by NJ, ML, and BI methods. In addition,
S. dermogenysi n. sp., originating from the freshwater fish
D. pusillus, was also separated from both Thai and Korean
S. falcatus isolates originating from brackish water mullets
P. subviridis and humans, respectively. The intraspecific distance (K2P) based on the
CO1 gene of the
Stellantchasmus group was around 5%, while the distance (K2P) between
S. dermogenysi n. sp. and 2
S. falcatus isolates averaged 19.9%. This distance was also high when compared with
Metagonimus spp., at 7.5% [
35]. This molecular evidence, along with different morphological characters, clearly supports the distinction of the
Stellantchasmus species. The present findings agree with Wongsawad and Wongsawad [
22], who reported that differences in the morphology, habitat, DNA fingerprint, and phylogenetic relationships (based on HAT-RAPD) strongly suggested that
Stellantchasmus from freshwater and brackish water habitats are different species.
The morphological and molecular characteristics obtained in this study allows us to confidently identify the parasite isolate originating from the freshwater fish D. pusillus, in Thailand, as a new species, S. dermogenysi n. sp. Although S. dermogenysi shares many morphological similarities with S. falcatus, the molecular evidence supports the distinction. The present findings will help a definitive diagnosis in humans, may affect treatment, prevention measures, and research into anti-parasitic agents.
ACKNOWLEDGMENTS
This research work was partially supported by Chiang Mai University, Chiang Mai, Thailand. We thank the Applied Technology for Biodiversity, Institute for Science and Technology; Applied Parasitology, Department of Biology, Faculty of Science; Economic Plant Genome Service Centre, Faculty of Science, Chiang Mai University, Thailand for the provision of instruments, facilities, and access to laboratories. This work was supported by National Research Council of Thailand to CW (grant no. 2559A10402051, 2016) and The Thailand Research Fund to NN (grant no. TRG5880053). Special thanks are extended to the Natural History Museum, London, and Senior Curator of Parasites and Vectors Division.
Fig. 1(A) Stellantchasmus falcatus adult fluke originating from the brackish water fish P. subviridis. Ventral view. (B) Stellantchasmus dermogenysi n. sp. originating from the freshwater fish D. pusillus.
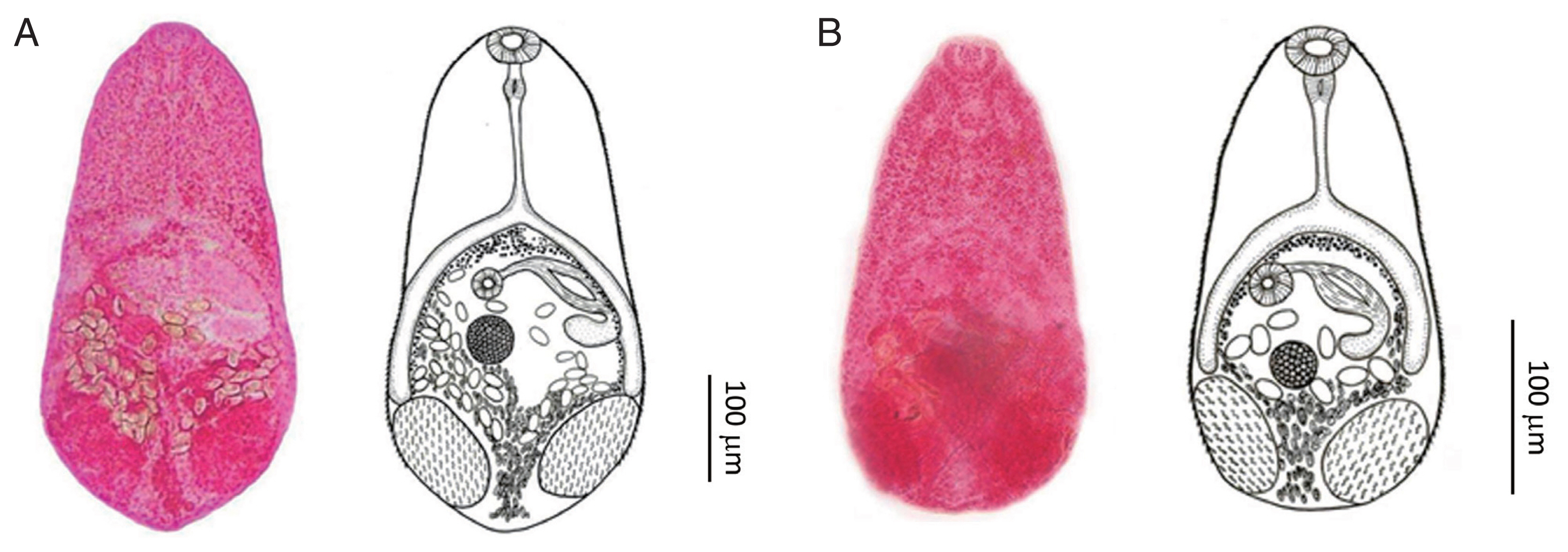
Fig. 2A phylogenetic tree of Stellantchasmus spp. and related group based on CO1 gene. Statistics can be referred to in Materials and Methods.

Table 1Helminth species used in this study and GenBank accession numbers for their corresponding CO1 sequence
Table 1
|
Species |
Host |
Country |
GenBank accession no. |
|
Stellantchasmus dermogenysi n.sp.a
|
Experimental hamster |
Chiang Mai, Thailand |
- |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753571 |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753572 |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753573 |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753574 |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753575 |
|
Stellantchasmus dermogenysi n.sp. |
Experimental hamster |
Chiang Mai, Thailand |
KU753576 |
|
Stellantchasmus falcatusa
|
Experimental hamster |
Chon Buri, Thailand |
AY382320 |
|
Stellantchasmus falcatusa
|
Korean human |
Korea |
- |
|
Stellantchasmus falcatus
|
Experimental hamster |
Chon Buri, Thailand |
KU753568 |
|
Stellantchasmus falcatus
|
Experimental hamster |
Chon Buri, Thailand |
KU753569 |
|
Stellantchasmus falcatus
|
Experimental hamster |
Chon Buri, Thailand |
KU753570 |
|
Metagonimus yokogawib
|
Experimental hamster |
Korea |
AB470519 |
|
Metagonimus takahashiib
|
Experimental hamster |
Korea |
AF096231 |
Table 2Comparison of body and organ measurements between Stellantchasmus falcatus and Stellantchasmus dermogenysi n. sp.
Table 2
|
Items |
Stellantchasmus falcatus
|
Stellantchasmus dermogen-ysi n. sp. from D. pusillus (B) |
Ratio A:B |
|
Onji & Nishio (1916) |
Pearson (1964) |
Seo et al. (1984) |
From a Korean humana
|
From P. subviridis (A) |
|
No. worms measured |
- |
10 |
10 |
5 |
27 |
30 |
|
|
Body length |
440 |
480 |
700 |
526 |
520 |
374* |
1.39 |
|
Body width |
220 |
270 |
360 |
326 |
222 |
167* |
1.33 |
|
Oral sucker length |
55 |
40 |
62 |
52 |
42 |
36 |
1.17 |
|
Oral sucker width |
55 |
46 |
61 |
51 |
48 |
42 |
1.14 |
|
Prepharynx length |
- |
Present |
- |
Present |
Present |
Almost absent |
- |
|
Pharynx length |
35 |
24 |
44 |
36.2 |
30 |
27 |
1.11 |
|
Pharynx width |
25 |
28 |
41 |
35.6 |
27 |
25 |
1.08 |
|
Esophagus length |
70–150 |
94 |
108 |
87.6 |
117 |
56b
|
2.09c
|
|
Ventral sucker length |
37 |
30 |
42 |
37 |
31 |
22b
|
1.41 |
|
Ventral sucker width |
37 |
29 |
42 |
37 |
31 |
24b
|
1.29 |
|
Expulsor length |
120 |
77 |
111 |
75 |
63 |
54 |
1.17 |
|
Expulsor width |
30 |
26 |
60 |
33 |
32 |
26 |
1.23 |
|
Ovary length |
50 |
46 |
71 |
65 |
54 |
53 |
1.02 |
|
Ovary width |
50 |
42 |
70 |
58.3 |
42 |
49 |
0.86c
|
|
Testis (R) length |
96 |
70 |
100 |
88.8 |
104 |
90 |
1.16 |
|
Testis (R) width |
78 |
57 |
71 |
85 |
56 |
58 |
0.97c
|
|
Testis (left) length |
88 |
70 |
96 |
102.5 |
105 |
91 |
1.15 |
|
Testis (left) width |
75 |
57 |
66 |
96.5 |
58 |
57 |
1.02 |
|
Egg (uterine) length |
27 |
23 |
27 |
27.1 |
21 |
21 |
1.00 |
|
Egg (uterine) width |
14 |
13 |
12 |
13.8 |
12 |
11b
|
1.09 |
Table 3Genetic distance of Stellantchasmus spp. based on CO1 gene
Table 3
|
Taxa |
S. falcatus
|
S. falcatusa
|
S. dermogenysi n. sp. |
|
S. falcatus
|
- |
0.014 |
0.024 |
|
S. falcatusa
|
0.083 |
- |
0.026 |
|
S. dermogenysi n. sp. |
0.194 |
0.217 |
- |
References
- 1. Pearson JC, Ow-Yang CK. New species of Haplorchis from Southeast Asia, together with keys to the Haplorchis-group of heterophyid trematodes of the region. Southeast Asian J Trop Med Public Health 1982;13:35-60.
- 2. Pearson JC. A revision of the subfamily Haplorchinae Looss, 1899 (Trematoda: Heterophyidae): I. The Haplorchis group. Parasitology 1964;54:601-676.
- 3. Onji Y, Nishio T. On intestinal distomes. Iji Shimbun 1916;949:589-593.
- 4. Seo BS, Hong ST, Chai JY, Lee SH. Studies on intestinal trematodes in Korea. VIII. A human case of Echinostoma hortense infection. Korean J Parasitol 1983;21:219-223.
- 5. Kliks M, Tantachamrun T. Heterophyid (trematoda) parasites of cats in North Thailand, with notes on a human case found at necropsy. Southeast Asian J Trop Med Public Health 1974;5:547-555.
- 6. Sohn WM, Chai JY, Lee SH. Two cases of natural human infection by Heterophyes nocens and the infection status of heterophyid metacercariae in mullets from Samcheonpo, Kyongnam Province. Inje Med J 1989;10:443-452.
- 7. Tantachamrun T, Kliks M. Heterophyid infection in human ileum: report of three cases. Southeast Asian J Trop Med Public Health 1978;9:228-231.
- 8. Sripalwit P, Wongsawad C, Chai JY, Anuntalabhochai S, Rojanapaibul A. Investigation of Stellantchasmus falcatus metacercariae in half-beaked fish, Dermogenus pusillus from four districts of Chiang Mai Province, Thailand. Southeast Asian J Trop Med Public Health 2003;34:281-285.
- 9. Africa CM, de Leon W, Garcia EY. Visceral complications in intestinal heterophyidiasis of man. Acta Med Phil (Monographic series) 1940;1:1-32.
- 10. Chai JY, Sohn WM. Identification of Stellantchasmus falcatus metacercariae encysted in mullets in Korea. Korean J Parasitol 1988;26:65-68.
- 11. Chuboon S, Wongsawad C. Molecular identification of larval trematodes in intermediate host from Chiang Mai, Thailand. Southeast Asian J Trop Med Public Health 2009;40:1216-1220.
- 12. Pubua J, Wongsawad C. Redescription of the trematode metacercariae from the mullet (Liza subviridis) and half-beak (Dermogenys pusillus). Southeast Asian J Trop Med Public Health 2007;38:106-109.
- 13. Noikong W, Wongsawad C, Chai JY, Saenphet S, Trudgett A. Molecular analysis of echinostome metacercariae from their second intermediate host found in a localised geographic region reveals genetic heterogeneity and possible cryptic speciation. PLoS Negl Trop Dis 2014;8:e2778.
- 14. Georgieva S, Selbach C, Faltýnková A, Soldánová M, Sures B, Skírnisson K, Kostadinova A. New cryptic species of the “revolutum” group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasit Vectors 2013;6:64.
- 15. Ho HW, Bray RA, Cutmore SC, Ward S, Cribb TH. Two new species of Phyllodistomum Braun, 1899 (Trematoda: Gorgoderidae Looss, 1899) from Great Barrier Reef fishes. Zootaxa 2014;3779:551-562.
- 16. Sereno-Uribe AL, Pinacho-Pinacho CD, García-Varela M, de León GPP. Using mitochondrial and ribosomal DNA sequences to test the taxonomic validity of Clinostomum complanatum Rudolphi, 1814 in fish-eating birds and freshwater fishes in Mexico, with the description of a new species. Parasitol Res 2013;112:2855-2870.
- 17. Nadler SA, DE León GP. Integrating molecular and morphological approaches for characterizing parasite cryptic species: implications for parasitology. Parasitology 2011;138:1688-1709.
- 18. Cable J, van Oosterhout C. The impact of parasites on the life history evolution of guppies (Poecilia reticulata): the effects of host size on parasite virulence. Int J Parasitol 2007;37:1449-1458.
- 19. Huyse T, Volckaert FA. Identification of a host-associated species complex using molecular and morphometric analyses, with the description of Gyrodactylus rugiensoides n. sp. (Gyrodactylidae, Monogenea). Int J Parasitol 2002;32:907-919.
- 20. Dyer NA, Ravel S, Choi KS, Darby AC, Causse S, Kapitano B, Hall MJ, Steen K, Lutumba P, Madinga J, Torr SJ, Okedi LM, Lehane MJ, Donnelly MJ. Cryptic diversity within the major trypanosomiasis vector Glossina fuscipes revealed by molecular markers. PLoS Negl Trop Dis 2011;5:e1266.
- 21. Wongsawad C, Rojanapaibul A, Vanittanakom P. Surface ultrastructure of encysted metacercariae and of adult Stellantchasmus sp. (Trematoda: Heterophyidae). J Electron Microscop Soc Thailand 1997;11:19-26.
- 22. Wongsawad C, Wongsawad P. Molecular markers for identification of Stellantchasmus falcatus and a phylogenic study using the HAT-RAPD method. Korean J Parasitol 2010;48:303-307.
- 23. Chontananarth T, Wongsawad C, Chomdej S, Krailas D, Chai JY. Molecular phylogeny of trematodes in Family Heterophyidae based on mitochondrial cytochrome c oxidase subunit I (mCOI). Asian Pac J Trop Med 2014;7:446-450.
- 24. Sripalwit P, Wongsawad C, Chontananarth T, Anuntalabhochai S, Wongsawad P, Chai JY. Developmental and phylogenetic characteristics of Stellantchasmus falcatus (Trematoda: Heterophyidae) from Thailand. Korean J Parasitol 2015;53:201-207.
- 25. Wongsawad C. Development of HAT-RAPD marker for detection of Stellantchasmus falcatus infection. Southeast Asian J Trop Med Public Health 2011;42:46-52.
- 26. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.
- 27. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 2012;9:772.
- 28. Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4. Sunderland, USA. Sinauer Associates; 2003.
- 29. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696-704.
- 30. Felsenstein J. Bootstraps and testing trees. [Internet]; Availabe from: http://evolution.gs.washington.edu/sisg/2016/2016_SISG_19_7.pdf
- 31. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 2010;39:783-791.
- 32. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-542.
- 33. Chai JY, Sohn WM, Na BK, Jeoung HG, Sinuon M, Socheat D. Stellantchasmus falcatus (Digenea: Heterophyidae) in Cambodia: discovery of metacercariae in mullets and recovery of adult flukes in an experimental hamster. Korean J Parasitol 2016;54:537-541.
- 34. Chai JY, Sohn WM, Na BK, Park JB, Jeoung HG, Hoang EH, Htoon TT, Tin HH. Zoonotic trematode metacercariae in fish from Yangon, Myanmar and their adults recovered from experimental animals. Korean J Parasitol 2017;55:631-641.
- 35. Pornruseetairatn S, Kino H, Shimazu T, Nawa Y, Scholz T, Ruangsittichai J, Saralamba NT, Thaenkham U. A molecular phylogeny of Asian species of the genus Metagonimus (Digenea)--small intestinal flukes--based on representative Japanese populations. Parasitol Res 2016;115:1123-1130.