Abstract
The endosymbionts of 4 strains of Acanthamoeba (KA/E9, KA/E21, KA/E22, and KA/E23) isolated from the infected corneas of Korean patients were characterized via orcein stain, transmission electron microscopic examination, and 16S rDNA sequence analysis. Double membrane-bound, rod-shaped endosymbionts were distributed randomly throughout both the trophozoites and cysts of each of Acanthamoeba isolates. The endosymbionts of KA/E9, KA/E22, and KA/E23 were surrounded by electron-translucent areas. No lacunae-like structures were observed in the endosymbionts of KA/E21, the bacterial cell walls of which were studded with host ribosomes. Comparative analyses of the 16S rDNA sequences showed that the endosymbionts of KA/E9, KA/E22 and KA/E23 were closely related to Caedibacter caryophilus, whereas the KA/E21 endosymbiont was assigned to the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum. In the 4 strains of Acanthamoeba, the hosts of the endosymbionts were identified as belonging to the Acanthamoeba castellanii complex, which corresponds to the T4 genotype. Acanthamoeba KA/E21 evidenced characteristics almost identical to those of KA/E6, with the exception of the existence of endosymbionts. The discovery of these endosymbionts from Acanthamoeba may prove essential to future studies focusing on interactions between the endosymbionts and the amoebic hosts.
-
Key words: Acanthamoeba, bacterial endosymbiont, keratitis, 16S rDNA, 18S rDNA
INTRODUCTION
The
Acanthamoeba genus is an opportunistic protozoan parasite which has adapted to live in a variety of environments, including soil, air, fresh water, ocean sediment, wild animals, and humans. Some
Acanthamoeba species have been determined to induce serious human infections, including amoebic keratitis, particularly in contact lens wearers, as well as granulomatous amoebic encephalitis (GAE) in immunocompromised individuals (
Marciano-Cabral and Cabral, 2003). Amebic keratitis was considered to be a relatively rare type of corneal infection during the 10 years following the first case report by Jones et al. (
1975). However, the incidence of amoebic keratitis infection has exponentially increased over the past 2 decades, as the consequence of the increasing use of contact lenses. Recently, the incidence of amoebic keratitis in Korea has also evidenced an increase; this may also be due to the high
Acanthamoeba contamination rate of contact lenses cases in Korea, as has been reported in several papers (
Lee et al., 1997;
Yu et al., 2001;
Kong et al., 2002).
Bacteria are a major food source for amoeba (
Rodriguez-Zaragoza, 1994). However, some bacteria are capable of surviving within
Acanthamoeba, and exploit them as hosts. Bacterial transient intracellular associations with
Acanthamoeba have been previously reported, and have included species such as
Legionella pneumophila,
Chlamydia pneumoniae,
Mycobacterium avium, and
Escherichia coli O157 (
Rowbotham, 1986;
Essig et al., 1997;
Steinert et al., 1998;
Barker et al., 1999). Within the amoebal trophozoites, these bacteria may be protected against adverse environmental conditions, thereby suggesting that amoebae may play a crucial role in the survival and dispersal of bacteria within the environment (
Winiecka-Krusnell and Linder, 2001). Such interactions may be of clinical relevance, particularly because the multiplication of
L. pneumophila and
M. avium within the amoebic host cell increases bacterial virulence (
Cirillo et al., 1997).
In this study, we describe a new discovery regarding the presence of endosymbionts in 4 strains of Acanthamoeba (KA/E9, KA/E21, KA/E22, KA/E23) isolated from the infected corneas of Korean patients. The amoebic host and endosymbiotic bacteria were characterized via transmission electron microscopy and 18S and 16S rDNA sequence analyses.
MATERIALS AND METHODS
Isolation and maintenance of Acanthamoeba
Four
Acanthamoeba isolates; KA/E9, KA/E21, KA/E22, and KA/E23, all of which harbored bacterial endosymbionts, were isolated from the infected corneas of Korean patients. Corneal samples collected by scraping from keratitis patients were loaded onto 1.5% agar plates covered with heat-inactivated
Escherichia coli (ATCC 25922, plasmid-free, Washington D.C., USA). Culturing on the plates, clonal isolation, and axenic culturing were conducted via the same techniques as previously described (
Yu et al. 2001).
Orcein staining was employed in order to visualize the presence of endosymbionts within the
Acanthamoeba, as described by Beier et al. (
2002). Thin films of
Acanthamoeba were prepared on slides and allowed to air-dry. The slides were then placed in 1% KMnO
4 for 5 min, washed in water, and rinsed for 30 sec in 3% oxalic acid, and finally rinsed off in distilled water. The films were covered with a few drops of orcein-carmine staining solution. The slides were examined 1 hr after staining under a light microscope (Olympus IX70, Tokyo, Japan), using the oil immersion objective.
The
Acanthamoeba MtDNA was extracted via the method described by Yagita and Endo (
1990).
Acanthamoebae trophozoites harvested at the end of the logarithmic growth phase were washed in cold PBS. The amoebas were lysed with fresh 1% sodium dodecyl sulfate (SDS) solution in 0.2 N NaOH and potassium acetate buffer, and incubated for 30 min on ice. The MtDNA was then extracted with phenol and phenol-chloroform (1:1) and recovered via precipitation with cold absolute ethanol in the presence of sodium acetate. The extracted MtDNA was then digested with the
BgL II restriction enzyme (Promega, Madison, Wisconsin, USA) at 37℃. The digested DNA was electrophoresed in 0.7% agarose gel stained with ethidium bromide. The MtDNA RFLP patterns were observed and photographed under a UV transilluminator.
The trophozoites were washed 3 times with cold PBS and prefixed for 2 hr with 4% glutaraldehyde in 0.1 M cacodylate buffer. After rinsing with 0.1 M cacodylate buffer, the sediment was post-fixed for 3 hr with 1% osmium tetroxide, rinsed twice with 0.1 M maleate buffer, pH 5.2, dehydrated with ethyl alcohol, and treated for 30 min with propylene oxide. The pellets were immersed overnight in propylene oxideresin (1:1) with continuous shaking, then embedded in resin and incubated overnight at 60℃. Ultrathin sections were cut on a Reichert-Jung ultramicrotome, and then stained with uranyl acetate and lead citrate. The sections were observed under a transmission electron microscope (Hitachi H-7000, Tokyo, Japan).
Sequence analysis of 16S rDNA of endo-symbionts and 18S rDNA of Acanthamoeba
In order to amplify the 16s rDNA of the bacterial endosymbionts, the MtDNA of each Acanthamoeba was employed as a template. The nucleotide sequences of the primers were as follows: forward; 5'-TCGACAACAGAGTTTGATCCTGGCTCAG-3' reverse; 5'-ATCCAAGCTTAAGGAGGTGATCC AGCC-3'. 16S rDNA amplification reactions were separately conducted in a thermal capillary cycler using a premix of 50 µl scale (iNtRON, Seongnam, Korea), which included 1.25 U of Taq polymerase (iNtRON). The PCR reactions were conducted at an initial 3 min at 94℃, followed by 30 cycles of 94℃ for 1.5 min, 50℃ for 1 min, 72℃ for 4 min, and a final 7-min elongation step at 72℃.
The chromosomal DNA of
Acanthamoeba isolates acquired via the method described by Kong and Chung (
1996) was utilized as a template for 18S rDNA PCR. The amplification of nearly full-length amoebic 18S rDNA gene fragments was conducted using primers originating from the ssu rRNA sequence of the
A. castellanii Neff strain, as reported by Gunderson and Sogin (
1986). The sequences of the primers were as follows: forward; 5'-CCGAATTCGTCGACAACCTGGTTGATCCTGCCAGT-3' reverse; 5'-GGATCCAAGCTTGATCCTTCTGCAGGTTCACCTAC-3'. The 18S rDNA amplification reactions were separately conducted in a thermal capillary cycler using premix of 50 µl scale (iNtRON). The PCR reactions were conducted for an initial 3 min at 94℃, followed by 30 cycles of 94℃ for 1 min, 58℃ for 1 min, 72℃ for 2.5 min, and a final elongation step of 10 min at 72℃. The presence and size of all of the amplified products were evaluated via agarose gel electrophoresis and ethidium bromide staining.
The amplified PCR products were ligated into the T/A cloning vector pGEM-T easy System I (Promega), with subsequent transformation into E. coli by each vector. DNA sequencing was conducted by an outside company (Macrogen, Seoul, Korea).
Phylogenetic analyses
Nucleotide sequences were compared to the sequences of published strains via BLAST search (
Altschul et al., 1990). CLUSTAL X (
Thompson et al., 1997) was used for pairwise alignment and calculations of the percentages of sequence dissimilarity. The percentages of similarity correspond to the number of sites that are identical between the 2 isolates, aligned with one another, as in the master alignment. Phylogeny was evaluated using the CLUSTAL X 1.82 program with a low gap penalty, and the phylogenetic trees were drawn using TreeView computer program.
RESULTS
Orcein staining
Bacterial endosymbionts were readily visualized within the orcein-stained
Acanthamoeba under phase-contrast light microscopy (
Fig. 1). Darkly stained rod-shaped bacterial endosymbionts were found to be distributed randomly within the cytoplasm of the trophozoites of
Acanthamoeba KA/E9, KA/E21, KA/E22, and KA/E23. In comparison, no bacteria were detected in
A. castellanii Castellani, which was employed as a control.
Fig. 2 shows the agarose gel electrophoretic patterns of
BgL II-digested MtDNA from 4 clinical
Acanthamoeba isolates and the reference strains. The restriction fragment patterns of the extranuclear circular DNA from
Acanthamoeba strains KA/E9, KA/E21, KA/E22, and KA/E23 evidenced extra fragments, as well as common fragments which were also observed in the reference strains.
Acanthamoeba KA/E9 and KA/E23 evidenced extremely similar MtDNA RFLP patterns with
A. triangularis SH621, whereas
Acanthamoeba KA/E21 and KA/E22 were identical to the MtDNA RFLP patterns of KA/E6 and KA/E2, respectively, obtained from other corneal isolates of Korean patients.
TEM
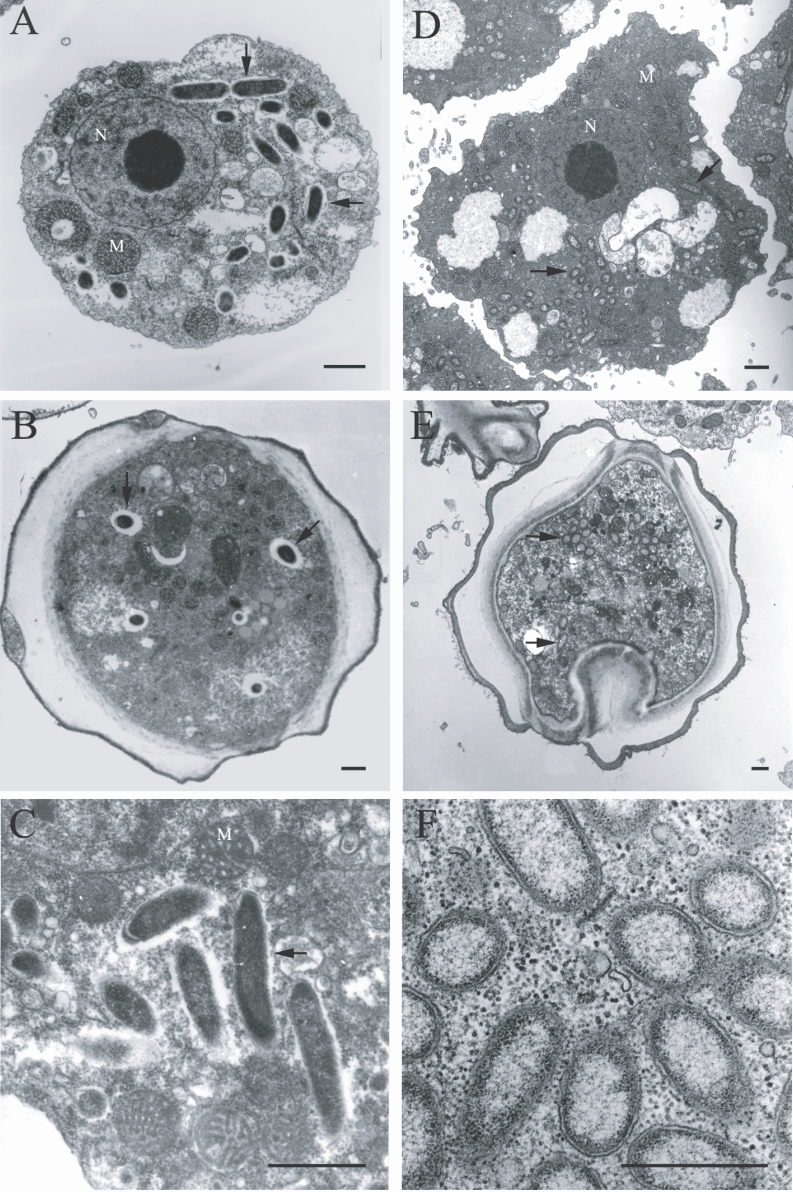
The morphological and ultrastructural characteristics of the endosymbionts were elucidated via transmission electron microscopy. The rod-shaped endosymbiont of
Acanthamoebae KA/E9 was estimated to be approximately 1.32 × 0.4 µm in size (
Fig. 3A-3C). In addition, the endosymbionts were determined to be randomly distributed within the host cytoplasm, and were frequently surrounded by an electron-translucent area (
Fig. 3C). The endosymbionts of
Acanthamoebae strains KA/E22 and KA/E23 were morphologically identical to those of KA/E9.
However, the endosymbionts of
Acanthamoeba KA/E21 were also determined to be randomly distributed within the cytoplasm of both the trophozoites and cysts (
Fig. 3D-3F). They appeared as straight rods with rounded ends, and measured approximately 1.08 × 0.35 µm. The endosymbionts were bound by double membranes, and the bacterial cell surfaces were studded with a number of host cell ribosomes (
Fig. 3F). They were not enclosed within vacuoles. No specific features could be observed within the bacteria in either case.
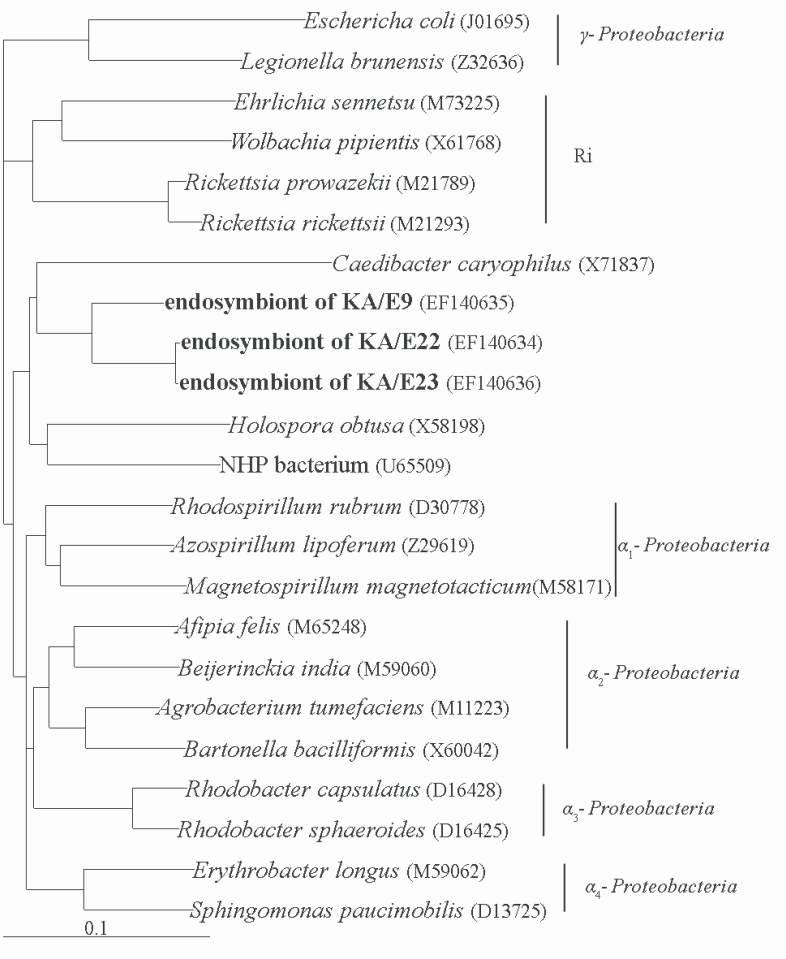
Comparative analysis with the 16S rRNA gene sequences showed that the endosymbionts of KA/E9 (84%), KA/E22 (83%) and KA/E23 (83%) evidenced a high degree of sequence similarity with
Caedibacter caryophilus, a member of the
α-Proteobacteria. Phylogenetic treeing methods showed that the endosymbionts of KA/E9 KA/E22 and KA/E23,
Caedibacter caryophilus,
Holospora obtusa and 'NHP bacterium' constitute a monophyletic cluster within the
α-Proteobacteria, and are closely related to the
Rickettsiaes (
Fig. 4).
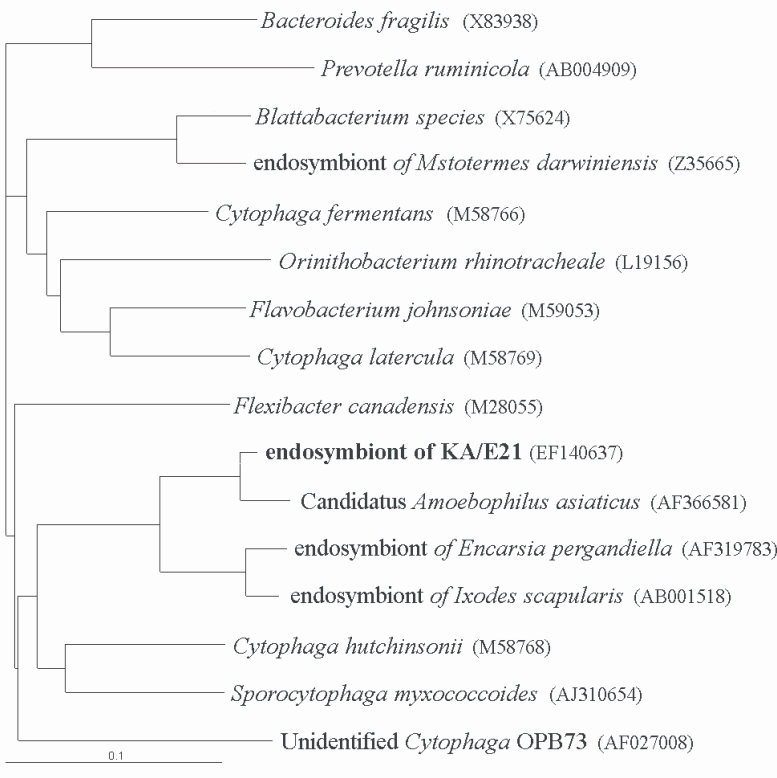
The results of comparative sequence analysis supported the assignment of the endosymbiont of
Acanthamoeba KA/E21 to the
Cytophaga-Flavobacterium-Bacteroides (CFB) phylum. The KA/E21 endosymbionts evidenced the highest degree of similarity, with 97% 16S rDNA sequence similarity to a recently published bacterium,
Candidatus Amoebophilus asiaticus (
Horn et al., 2001). This symbiont also evidenced a high degree of similarity with the endosymbionts of the tick,
Ixodes scapularis (90%) and with those of the parasitic white fly,
Encarsia pergandiella (89%). However, the endosymbiont of KA/E21 evidenced some similarity (less than 80%) to other species within the CFB phylum. The results of our phylogenetic analyses consistently demonstrated that the endosymbiont of KA/E21,
Candidatus Amoebophilus asiaticus, the endosymbiont of
I. scapularis, and the endosymbiont of
E. pergandiella constitute a monophyletic lineage within the CFB phylum (
Fig. 5).
Comparative sequence analysis verified the morphology-based identification of this isolate as
Acanthamoeba sp.
Acanthamoebae KA/E9, and KA/E23 evidenced the most pronounced sequence similarity (98%) with
A. triangularis SH621. In addition,
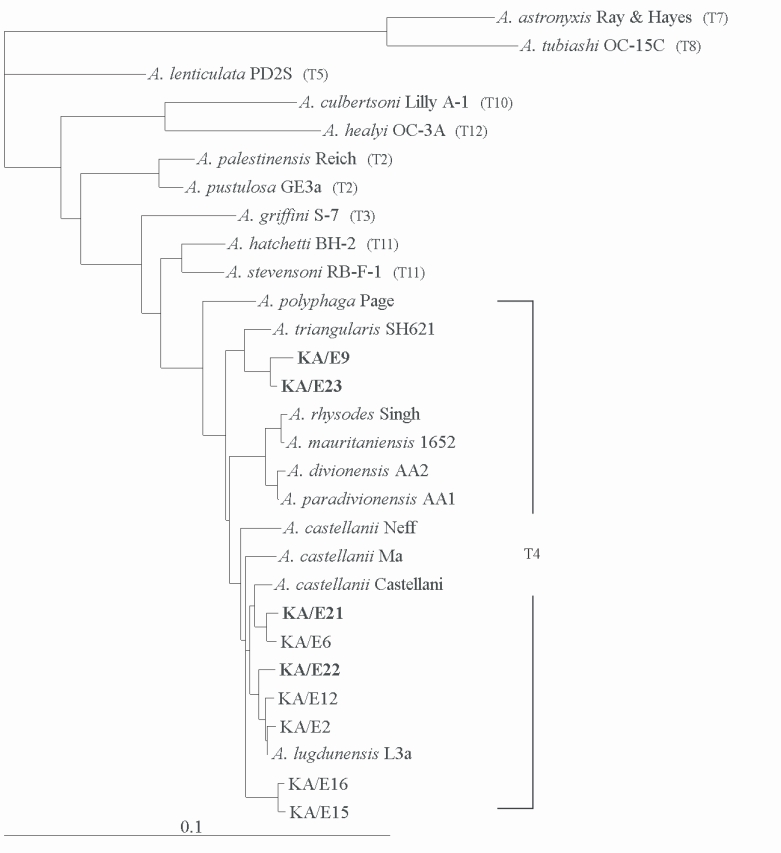
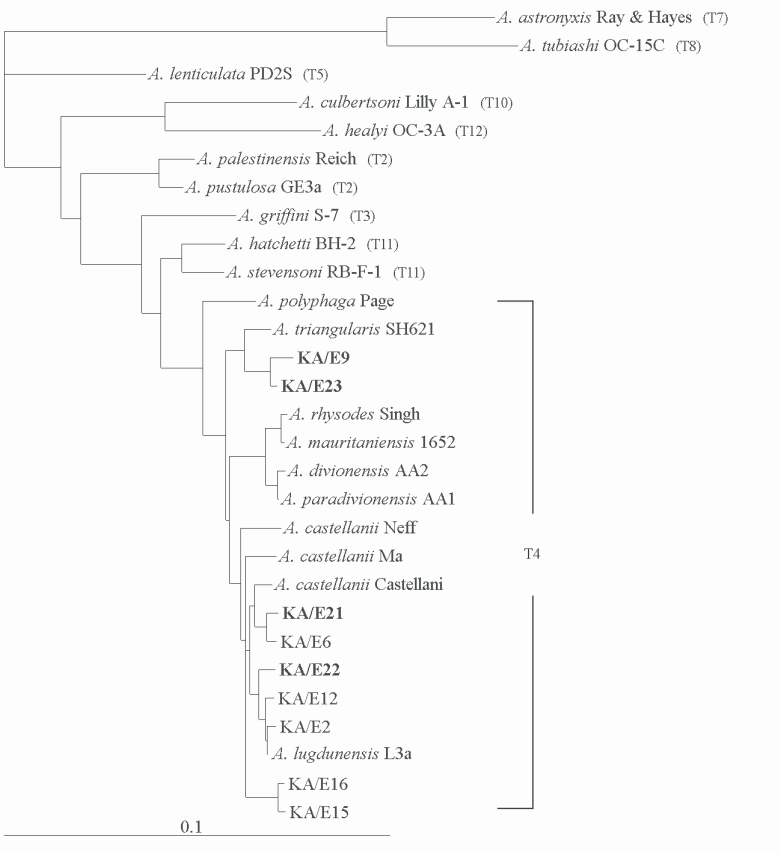
Acanthamoeba KA/E21 and KA/E22 evidenced very high 18S rDNA sequence similarity (99%) with the clinical isolates KA/E6 and KA/E2 (isolated from Korean keratitis patients), respectively. The dendrogram constructed on the basis of the 18S rDNA sequence analyses is shown in
Fig. 6. Using the 95% similarity value for the definition of the
Acanthamoeba 18S rDNA sequence types (
Stothard et al., 1998), the
Acanthamoeba host should be considered a constituent of the
A. castellanii complex, which corresponds to genotype T4, the genotype encompassing the majority of clinical and environmental isolates of
Acanthamoeba.
DISCUSSION
Bacterial endosymbionts are commonly observed in protozoans (
Winiecka-Krusnell and Linder, 2001). Approximately 24% of
Acanthamoeba, isolates from both environmental and medical samples were determined to harbor endosymbionts (
Fritsche et al., 1993). In this study, we have investigated, for the first time, the presence of endosymbiont bacteria in clinical
Acanthamoeba isolates from infected corneas of Korean patients.
Four
Acanthamoeba isolates suspected to harbor endosymbionts evidenced very peculiar MtDNA RFLP patterns. The estimated size of the MtDNA of these isolates, based on the numbers and sizes of the DNA fragments generated by RFLP, was much larger than the general size of
Acanthamoeba MtDNA reported by Burger et al. (
1995). These results might indicate that the 4 isolates of
Acanthamoeba may possibly harbor intracellular endosymbiotic bacteria containing circular DNAs in
Acanthamoeba.
The KA/E9, KA/E22, and KA/E23 endosymbionts were determined to be surrounded by electron-translucent areas in the TEM examination, a result similar to that reported by Horn et al. (
1999). Hall and Voelz (
1985) previously demonstrated that this translucent area was the consequence of the presence of bacterial capsular material. With regard to the endosymbionts of KA/E21, our results indicated that they were surrounded by a double membrane, which was decorated with numerous ribosomes of the
Acanthamoeba host. These morphological characteristics are very consistent with what has been observed with the endosymbionts of
Acanthamoeba isolated previously from contact lens storage cases in Korea (
Chung et al., 1997).
On the basis of the results of comparative 16S rDNA sequence analyses, the endosymbionts of KA/E9, KA/E22 and KA/E23 are members of the α-subclass of
Proteobacteria, and are clearly affiliated with a monophyletic cluster of intracellular bacteria that are distinct from, but related to, the
Rickettsiaes. The
α-Proteobacteria cluster comprises a number of as-yet uncultured microorganisms that appear to be capable of infecting distantly-related eukaryotes, including
Paramecium caudatum. Within this cluster, the KA/E9, KA/E22, and KA/E23 endosymbionts evidence sequence similarities of 84%, 83%, and 83%, respectively, to
C. caryophilus. Because Stackebrandt and Goebel (
1994) mentioned that ≥ 97% 16S rDNA sequence similarity refers to one species, the endosymbionts of KA/E9, KA/E22, and KA/E23, which evidence less than 97% sequence similarity, might be representative of a species other than
C. caryophilus.
The 18s rDNA sequence analyses of the 2 hosts, KA/E9 and KA/E23, evidenced a 99% similarity with each other, and both were quite closely related to A. triangularis SH621. The 16S rDNA sequence analysis of the endosymbionts of KA/E9 and KA/E23 also resulted in a finding of 92% similarity, and were identical to the endosymbionts of C. cariophilus. On the basis of this finding, it is possible to suggest that a particular endosymbiont may exhibit a preference to certain specific host characteristics.
The effects of bacteria on amoebic hosts are less well known. The external bacteria enhance the growth and binding of
Acanthamoeba trophozoites to contact lenses, and also enhance the survival of amoeba in biocidal lens care solution (
Cengiz et al., 2000). However, the mechanisms by which bacteria affect the virulence and pathogenicity of
Acanthamoeba infections have yet to be thoroughly elucidated. With the exception of the presence of endosymbionts,
Acanthamoeba KA/E9 and KA/E23 with
A. triangularis SH621,
Acanthamoeba KA/E21 with KA/E6, and
Acanthamoeba KA/E22 with KA/E2 were found to be closely phylogenetically related, on the basis of MtDNA RFLP and 18S rDNA sequence analyses. This isogenetic pair can be employed in studies of the relationship between the bacteria with their host,
Acanthamoeba. Further studies will focus on the roles of these endosymbionts that augment amoebic cytopathogenicity.
Notes
-
This research was supported by Kyungpook National University Research Team Fund, 2002.
This work was supported by the Brain Korea 21 Project in 2006.
References
Fig. 1Phase-contrast micrographs of endosymbionts of Acanthamoeba stained with orcein-carmine. Numerous bacterial symbionts in the cytoplasm of trophozoites of Acanthamoeba KA/E9, KA/E21, KA/E22, and KA/E23 are visible. A. castellanii Castellani is used as a control. Bar = 2 µm.

Fig. 2Agarose gel electrophoretic restriction fragment patterns of Bgl II-digested mitochondrial DNA of host Acanthamoeba spp. and the reference strains. Lane 1; KA/E9, 2; KA/E23, 3; A. triangularis SH621, 4; KA/E21, 5; KA/E6, 6; KA/E22, 7; KA/E2. M; Hind III digested lamda phage DNA as DNA molecular size standard. Acanthamoeba KA/E9, KA/E21, KA/E22, KA/E23 show extra bands (arrow heads) as well as common bands with the other reference strains.
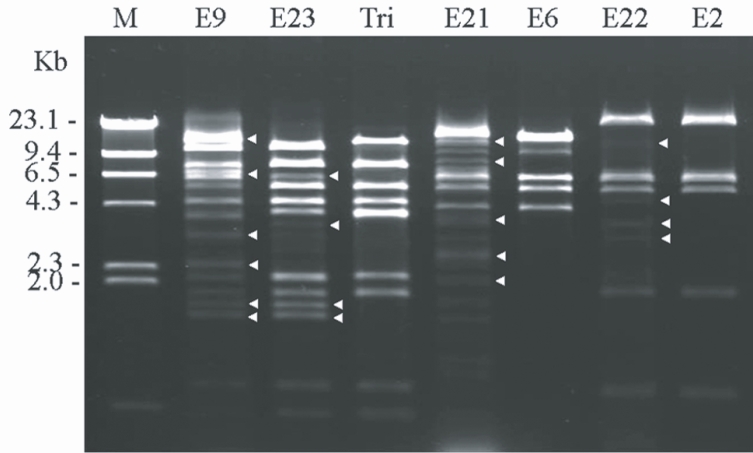
Fig. 3Transmission electron micrographs of Acanthamoeba KA/E9 (A-C) and KA/E21 (D-F) with bacterial endosymbionts. A, D: Trophozoites with numerous rod-shaped endosymbionts (arrows) in the cytoplasm. B, E: Cysts with endosymbionts. C: Ultrastructural feature of the bacteria surrounded by an eletron-translucent area. F: Bacterial endosymbionts closely associated with host ribosomes. N, nucleus; M, mitochondria. Bar = 1 µm.
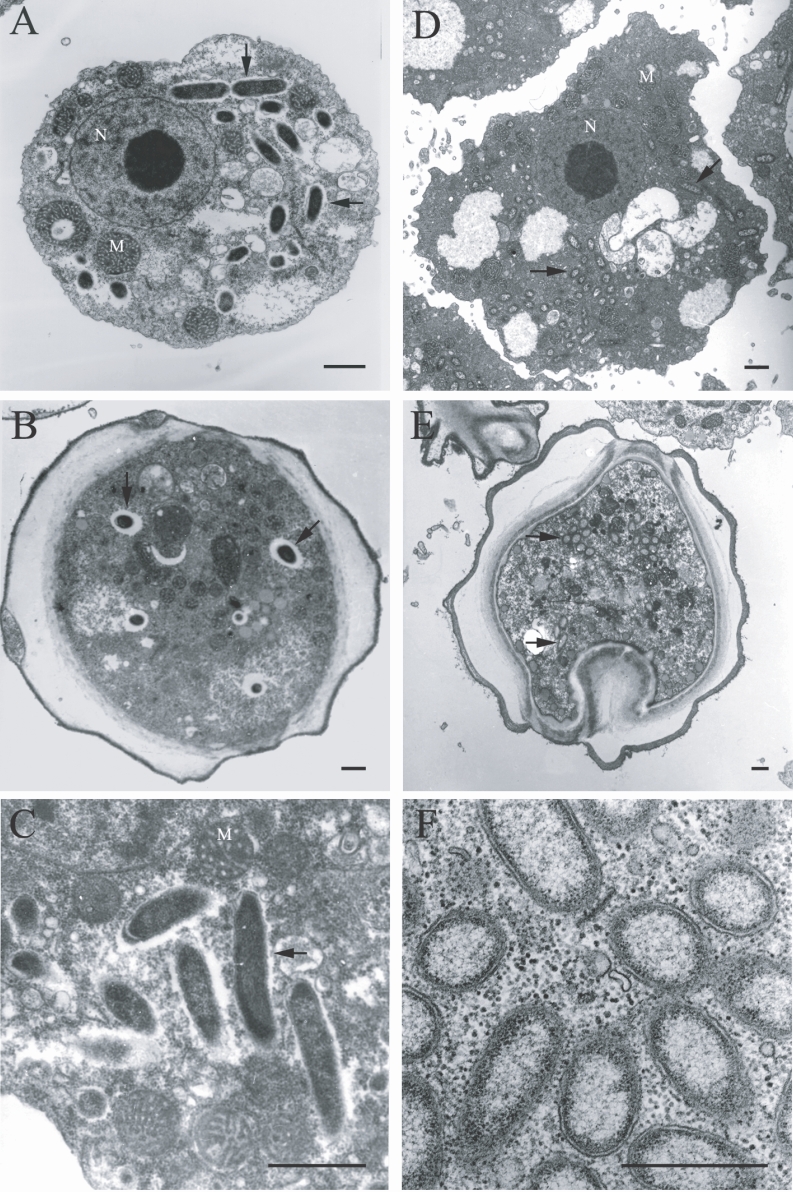
Fig. 4Phylogenetic tree, based on comparative 16S rDNA analysis, showing relationships of endosymbiont of Acanthamoeba KA/E9, KA/E22 and KA/E23 to representative members of the α-subclass of Proteobacteria.
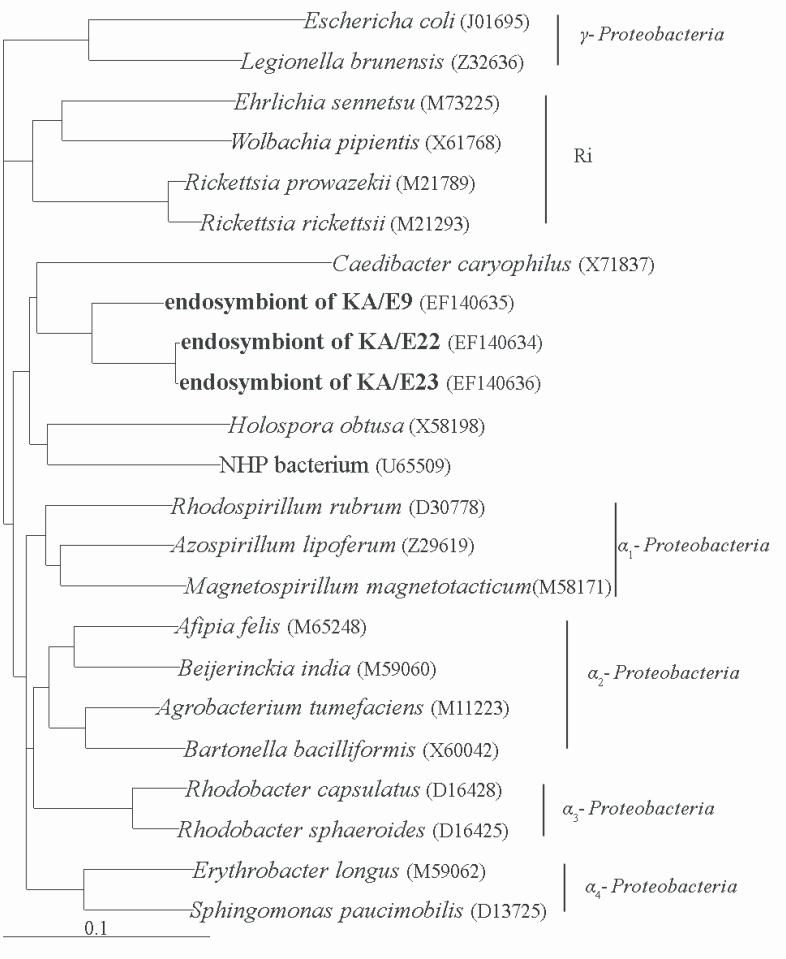
Fig. 5Phylogenetic tree, based on comparative 16S rDNA analysis, showing relationships of endosymbiont of Acanthamoeba KA/E21 to representative members of the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum.
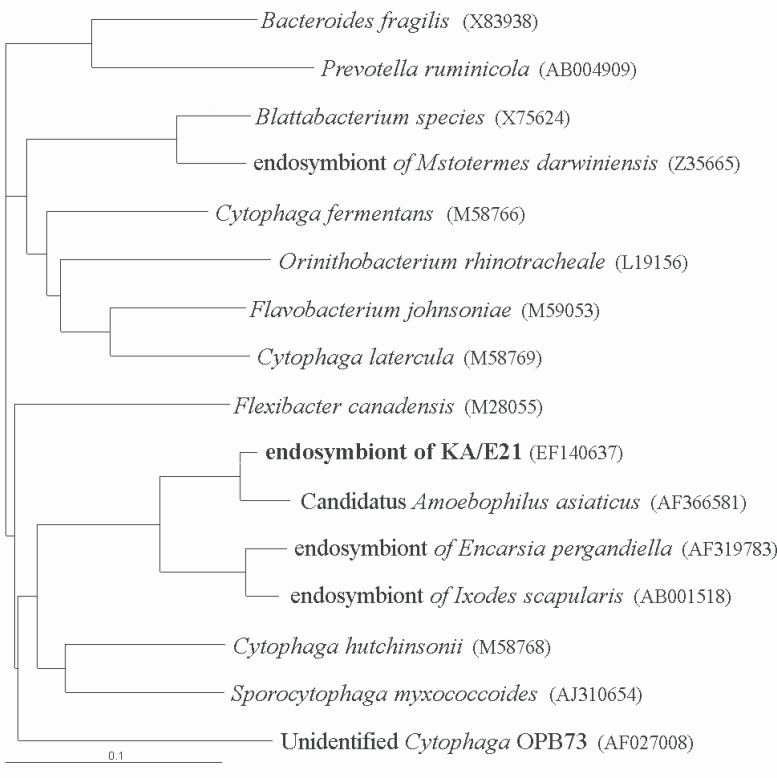
Fig. 618S rRNA-based Phylogenetic tree reflecting the affiliation of host Acanthamoeba sp. with selected members of genus Acanthamoeba. The reference with their corresponding accession numbers are: A. astronyxis Ray & Hayes, AF019064; A. tubiash OC-15C, AF019065; A. lenticulata PD2S, U94741; A. culbertsoni Lilly A-1, AF019067; A. healyi OC-3A, AF019070; A. palestinensis Reich, U07411; A. pustulosa GE3a, AF019050; A. griffini S-7, U07412; A. hatchetti BH-2, AF019068; A. stevensoni RB-F-1, AF019069; A. polyphaga Page, AF019061; A. triangularis SH621, AF316547; A. rhysodes Singh, AF351644; A. mauritaniensis 1652, AY351647; A. divionensis AA2, AY351646; A. paradivionensis AA1, AY351645; A. castellanii Castellani, U07413; A. lugdunensis L3a, AF005995; A. castellanii Neff, AF690457; A. castellanii Ma, U07414; strain KA/E2, AF005998; KA/E6, AF349044; KA/E9, AF316546; KA/E12, AF316545; KA/E15, AY148961; KA/E16, AY148962; KA/E21, EF140633; KA/E22, EF140638; KA/E23, EF140625.