Molecular Surveillance of Pfkelch13 and Pfmdr1 Mutations in Plasmodium falciparum Isolates from Southern Thailand
Article information
Abstract
Artemisinin-based combination therapy (ACT) resistance is widespread throughout the Greater Mekong Subregion. This raises concern over the antimalarial treatment in Thailand since it shares borders with Cambodia, Laos, and Myanmar where high ACT failure rates were reported. It is crucial to have information about the spread of ACT resistance for efficient planning and treatment. This study was to identify the molecular markers for antimalarial drug resistance: Pfkelch13 and Pfmdr1 mutations from 5 provinces of southern Thailand, from 2012 to 2017, of which 2 provinces on the Thai- Myanmar border (Chumphon and Ranong), one on Thai-Malaysia border (Yala) and 2 from non-border provinces (Phang Nga and Surat Thani). The results showed that C580Y mutation of Pfkelch13 was found mainly in the province on the Thai-Myanmar border. No mutations in the PfKelch13 gene were found in Surat Thani and Yala. The Pfmdr1 gene isolated from the Thai-Malaysia border was a different pattern from those found in other areas (100% N86Y) whereas wild type strain was present in Phang Nga. Our study indicated that the molecular markers of artemisinin resistance were spread in the provinces bordering along the Thai-Myanmar, and the pattern of Pfmdr1 mutations from the areas along the international border of Thailand differed from those of the non-border provinces. The information of the molecular markers from this study highlighted the recent spread of artemisinin resistant parasites from the endemic area, and the data will be useful for optimizing antimalarial treatment based on regional differences.
INTRODUCTION
Malaria remains a leading health problem in many countries, particularly those in tropical and subtropical regions of the world [1]. The countries of Greater Mekong Subregion (GMS) (i.e., Thailand, Cambodia, Laos, Myanmar, Vietnam, and China) have developed resistance to antimalarial drugs [2] although they set the target to eliminate malaria by 2030 [3]. In the late 1950s, the chloroquine resistance was initially reported in the Cambodia–Thailand border. It was then followed by the resistance to sulfadoxine-pyrimethamine, mefloquine and quinine [4]. Artemisinin-based combination therapy (ACT), either with mefloquine, lumefantrine, or piperaquine, is adopted as the main line of treatment for uncomplicated falciparum malaria [5]. The artemisinin kills the asexual blood stage of the parasites and it rapidly clears parasites than any other antimalarial drugs could do [6,7], while the partner drug eliminates the remaining parasites [8]. However, in 2006, the first observed reduction of parasites clearance was detected in patients from western Cambodia after an artemisinin therapy [9]. These cases confirmed that the artemisinin resistance had occurred along the Cambodia–Thailand border, the same area where chloroquine resistance was observed several decades ago [5,10]. Recently, the artemisinin resistance has already spread to 5 countries of the GMS [11]. Unfortunately, there is a simultaneous emergence of partner drug resistance, including resistance to artemether-lumefantrine, artesunate-amodiaquine, artesunate-mefloquine and artesunate-sulfadoxine-pyrimethamine, resulting in high treatment failure rate along Cambodian-Thai, Cambodia-Laos, and Thai-Myanmar borders [12].
Mutations in PfKelch13 propeller region were usually associated with artemisinin-resistance [13] and the highly frequent mutations detected in the eastern GMS (Cambodia, Lao PDR, and Viet Nam) were C580Y, R539T, Y493H, and I543T. On the other hand, F446L, N458Y, P574L, and R561H mutations were common in the western GMS (China, Myanmar, and Thailand) [14]. Non-synonymous (NS) mutations in PfKelch13 gene such as Y493H, R539T, I543T, and C580Y are the established molecular markers for artemisinin resistance in P. falciparum [15]. The C580Y mutation was common in western Cambodia, north-eastern Thailand, and southern Laos. Mutations in P. falciparum multidrug resistance gene (Pfmdr1) caused different in vivo efficacy and in vitro P. falciparum susceptibility to ACT including amodiaquine [16,17], mefloquine [18,19], lumefantrine [20,21] and artemisinin derivatives [19]. It was suggested that changes in sequence or copy number of Pfmdr1 altered influx and efflux of several drugs in the parasite food vacuole [22]. The 5 common genotypes of Pfmdr1 were N86Y, Y184F, S1034C, N1042D, and D1246Y. Pfmdr1 N86Y mutation was associated with chloroquine and amodiaquine resistance [2–4], while Y184F mutation was common with increased resistance to mefloquine and artesunate [23]. Point mutations at S1034C, N1042D, and D1246Y have been reported as resistance to quinine and increased susceptibility to mefloquine, halofantrine, and artemisinin derivatives [19,24,25].
Southern Thailand consists of 14 provinces with a population of about 8.7 million. It is in borders with Malaysia to the south and Myanmar to its upper west. In Malaysia, the incidence of malaria is estimated to be 13 cases per 100,000 populations [26] and the prevalence of antimalarial drug resistance was relatively low [27]. The molecular analysis showed a single copy of Pfmdr1 and wild-type PfKelch13 genes indicating that Malaysia has limited case of artemisinin resistance. However, the prevalence of malaria is higher in southern Thailand especially in areas bordering with the countries such as Myanmar. The dramatic increase of malaria cases in southern Thailand is attributed to its tropical rainforest, relatively higher proportion of migrant workers for rubber plantations from neighboring countries, and the prolonged insurgency in some provinces [4].
The aim of the present study is to analyze the molecular markers associated with P. falciparum resistance to antimalarial drugs in the Kelch13 and Pfmdr1 genes from the samples collected from Southern Thailand. The results would provide crucial evidence for further molecular surveillance of ACT in this part of the world. The information from this study can be used for efficient planning and targeting of malaria control in southern Thailand.
MATERIALS AND METHODS
Blood collection
The dried blood spots samples were received from the Office of Disease Prevention and Control 11 and 12, Thailand. The samples were collected from uncomplicated falciparum malaria patients when they attended their first visit to the malaria clinic and were positive on microscopic examination, but they did not have prior antimalarial treatment. Neither treatment outcome nor parasite clearance was followed up. The blood samples were obtained by finger-prick and spotted on filter paper (Whatman 3MM, GE Healthcare, Buckinghamshire, UK) for approximately 80 μl. Individual samples were stored in each plastic bag at room temperature before DNA extraction. DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Genomic DNA was eluted into 100 μl of elution buffer and used immediately or stored at −20°C until use.
Ethical approval for this study was obtained from the Faculty of Medicine, Prince of Songkla University (REC60-096-19-2).
Detection of PfKelch13 propeller gene
The presence of P. falciparum was confirmed in all samples by nested PCR with species-specific primers based on 18s rRNA gene, as described by Snounou et al. [28]. P. falciparum kelch13 was amplified from the genomic DNA using nested PCR following the protocol previously described by Ariey et al. [13] with some modifications. In the first PCR, the full-length Pfkelch13 gene sequence (1–2,283 bp) was amplified with a primer pair K13-1F (5′-TGGAAGGAGAAAAAGTAAAAACAAAA-3′) and K13-2283R (5′-TGTGCATGAAAATAAATATTAAAGAAG-3′). The PCR product was then amplified into 3 fragments in the second PCR step. The length of fragment 1 was 718 bp using K13-1F (see above) and K13c694-R (5′-TCTCGAATAAAATTCATTTGTGTCTT-3′). The fragment 2 covered positions 621-1538 using the primers K13c621F (5′-CGGAATTAAGTGATGCTAGTGA-3′) and K13c1538R (5′-CGATCATACACCTCAGTTTCAA-3′) to produce 917 bp product. The third fragment was amplified at the nucleotide positions 1344-2129 with the primer pairs K13c1344F (5′-AGGTGGATTTGATGGTGTAGAA-3′) and K13c2129R (5′-GGGCCAAGCTGCCATTCATTCATTTGT-3′) to produce amplicons of 786 bp. In each PCR reaction, 5 μl of genomic DNA was amplified in 25 μl of PCR master mix using 2 mM MgCl2 for first round PCR and 3 mM MgCl2 for second round PCR, 250 μM of dNTPs, 250 nM of oligonucleotide primers, 0.5 units of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, California, USA), and 3 μl of PCR product from the first PCR for the second PCR. The first PCR was performed with an initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, extension at 72°C for 2 min and the final extension at 72°C for 10 min. The second PCR was done with 35 cycles of denaturation at 94°C for 1 min, annealing for 2 min at 55°C for fragment 1 and 58°C for fragment 2, extension at 72°C for 2 min, and the final extension at 72°C for 5 min. The PCR products were analyzed on a 2% agarose gel electrophoresis stained with 0.5 μg/ml ethidium bromide and visualized under UV light.
Detection of Pfmdr1 gene
Five mutations of the Pfmdr1 gene were amplified using the 2 primer pairs as described in the previous study [29]. The sequences of codons 86 and 184 were amplified with primer pairs MDR1F (5′-AGAGAAAAAAGATGGTAACCTCAG-3′) and MDR1R (5′-ACCACAAACATAAATTAACGG-3′) with an expected amplified amplicons length of 590 bp. The second amplicon was amplified with the primer pairs MDR2F (5′-GCGGAGTTTTTGCATTTAGTTCAGATGATG-3′) and MDR2R (5′-AGCAGCAAACTTACTAACACGTTTAACATC-3′) to have the fragment length of 968 bp which contained the sequences of codons 1034, 1042, and 1246. The total reaction mixture of 25 μl consisted 1×PCR buffer, 3 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphate (dNTP), 0.2 μM of each oligonucleotide primers, 0.5 units of Platinum Taq polymerase and 5 μl of DNA template. The target amplification of MDR1 was programmed for initial denaturation at 95°C for 5 min, 35 cycles of 95°C for 1 min, 55°C for 1.30 min, 72°C for 2 min and final extension at 72°C for 8 min. For MDR2F/R, the conditions were an initial denaturation at 95°C for 3 min followed by 35 cycles of 93°C for 30 sec, 53°C for 30 sec, 72°C for 1 min and final extension at 72°C for 5 min. The PCR products were evaluated by electrophoresis on a 1.5% agarose gel and visualized with ethidium bromide staining.
Sequence analysis
The PCR products of PfKelch13 and Pfmdr1 amplicons were cleaned using QIAquick PCR purification kit (Qiagen) before sequencing by ABI Sequencer (Macrogen, Seoul, Korea). DNA sequences were aligned using BioEdit v7.2.5 with the reference sequences PF3D7_1343700 for PfKelch13 and PF3D7_0523000 for Pfmdr1 respectively.
PCR or sequencing was repeated if the amplification was unsuccessful, or the sequence chromatogram was vague at some point. In addition, some of the samples were repeated for sequencing to confirm the result. Data analysis was performed using Microsoft Excel 2007 and analyzed by SPSS 19.0 (SPSS Inc., Chicago, Illinois, USA).
RESULTS
A total of 128 uncomplicated falciparum malaria samples were obtained from 5 provinces of Southern Thailand (Fig. 1) from 2012 to 2017. All the samples were divided into 2 groups according to their geographical locations. The first group was from the 3 provinces that shared a border with the neighboring countries of Thailand and the second group was from the 2 provinces which were not located at the Thai border. The first group consisted of 16 samples from Chumphon (10 samples in 2013, 6 samples in 2014), 40 samples from Ranong (8 samples in 2012, 13 samples in 2013, 19 samples in 2015) and 12 samples from Yala in 2017 respectively. Chumphon and Ranong provinces are located at the Thai-Myanmar border while Yala shares border with Malaysia. The second group had 60 samples which were from Phang Nga and Surat Thani provinces and comprised of 14 samples from Phang Nga (4 samples in 2012, 10 samples in 2015), and 46 samples from Surat Thani (17 samples in 2014, 19 samples in 2015, 10 samples in 2016). Table 1 summerises the molecular analysis of PfKelch13 and Pfmdr1 genes together with the respective year of sample collection.
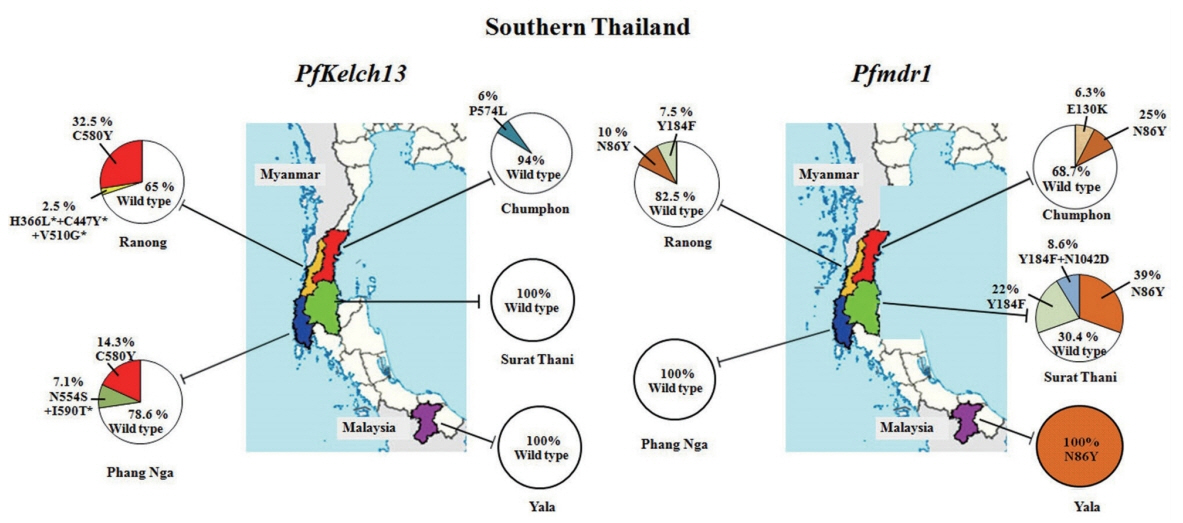
Geographic distribution of P. falciparum Kelch13 and Pfmdr1 mutations collected in 5 provinces of Southern Thailand including 2 provinces on the Thai-Myanmar border (Chumphon and Ranong), 2 from non-border provinces (Surat Thani and Phang Nga) and 1 from Thai-Malaysia border (Yala). Pie charts show the percentages of the mutations in 2 genes observed among 128 parasite samples collected between 2012 and 2017.

Summary of PfKelch13 and Pfmdr1 mutations identified in 5 southern provinces of Thailand together with the year of sample collection
PfKelch13 mutations
In this study, artemisinin resistance-confirmed mutations, C580Y and P574L were identified in provinces located at Thai-Myanmar border, Chumphon and Ranong, and in one non-border province, Phang Nga. C580Y mutation was detected in 13 of 40 samples (32.5%) from Ranong: 1 of 8 samples (12.5%) in 2012 and 6 of 13 samples (46.15%) in 2013 and 6 of 19 samples (31.58%) in 2014, respectively. It is interesting to note that the samples from Phang Nga, only 230 km south of Ranong, showed C580Y mutation in 2 of 10 samples (20%) in 2015. Chumphon province had P574L mutation in 1 of 6 samples (16.67%) in 2014, and 10 samples received in 2013 had no PfKelch13 mutation. Two samples had more than one mutations in PfKelch13 gene from the samples of the same year 2012. Of these, one isolate from Ranong had the mutation at N554S/I590T, and the other isolate (1 of 4 samples) from Phang Nga had H366L+C447Y+V510G, the 3 mutations present in a single sample which had not been detected elsewhere. Of these mutations, N554S has been reported from Mali in 2011 [30] and Ungoye, Kenya, in 2012 [31]. Unlike themutation of PfKelch13 presented on Thai-Myanmar border, P. falciparum circulated in Thai-Malaysia border, Yala in 2017, had 100% wild-type on PfKelch13 polymorphism. Forty-six isolates in 2014–2016 from Surat Thani, the non-border province had no evidence of molecular markers associated with artemisinin resistance.
Pfmdr1 mutations
Out of 128 samples, 72 samples (56.25%) showed the Pfmdr1 wild-type while 38 samples (29.69%) had N86Y mutation. Less frequently, E130K was observed in 1 (0.78%), Y184F in 13 (10.16%), and Y184F plus N1042D in 4 (3.12%) samples respectively. However, mutations at S1034C and D1246Y were not identified in the present study. Two provinces along the Thai Myanmar border, Chumphon and Ranong had the N86Y mutation in 20% (2/10 samples) in 2013, 33.33% (2/6 samples) in 2014, 23.08% (3/13 samples) in 2013, and 5.26% (1/19 samples) in 2015, respectively whereas Pfmdr1 Y184F mutation was detected in 7.69% (1/13 samples) in 2013 and 10.53% (2/19 samples) in 2015, but this mutation was not found in Chumphon. E130K was found in one isolated from Chumphon in 2014. This study was the first report of Pfmdr1 isolated from parasites in Surat Thani province. The N86Y mutation was observed in 7/17 samples (41.18%) in 2014, 4/19 samples (21.05%) in 2015 and 7/10 samples (70%) in 2016 respectively. For Y184F mutation, it was detected for 17.6% in 2014 (3/17 samples), 26.3% in 2015 (5/19 samples) and 20% in 2016 (2/10 samples) respectively. Double mutant genotypes Y184F plus N1042D appeared in the present study for 15.7% in 2015 (3/19 samples) and 10% in 2016 (1/10 samples). No mutations on Pfmdr1 gene associated with multidrug resistance were identified in the parasites isolated from Phang Nga province. However, in Yala province (Thai-Malaysia border), the point mutation N86Y in Pfmdr1 was present in 100% of the isolates.
DISCUSSION
Several studies have reported the mutations conferring drug resistance was commonly present in the area such as Thai-Cambodian, Thai-Myanmar and Thai-Laos borders. Drug resistance in malaria is frequently checked using molecular markers such as PfKelch13 gene for Artemisinin resistance, and P. falciparum multidrug resistance1 gene, for the partner drugs resistance such as chloroquine mefloquine, lumefantrine and quinine [32]. Since the last decade, the increasing prevalence of confirmed mutations for artemisinin resistance in PfKelch13 have been reported along the Thai-Cambodia and Thai-Myanmar borders, and in the GMS region [13,14,33–35]. The people with subclinical infection carrying the resistant P. falciparum allele might contribute to the spread of artemisinin-resistant gene to Thailand via cross-border movements. Mae Hong Son, one of the northwestern provinces, and Surat Thani and Yala provinces in southern Thailand have limited artemisinin-resistant genes [36] even though they are in the endemic area and are mainly rural, forested and bordering with Myanmar. Hence, it is crucial to monitor the current status of artemisinin resistance in the region using molecular markers as well as therapeutic efficacy, especially in an area where we have limited knowledge for P. falciparum drug resistance.
Since 2009, the first-line treatment for uncomplicated P. falciparum infections had been changed from 2-days artesunate-plus-mefloquine to 3-days regimen in eastern and western Thailand [5,15,37]. However, widespread mefloquine resistance has been reported in Thailand because it was used as monotherapy over the last few decades. Data from the present study revealed the PfKelch13 and Pfmdr1 mutations from 5 Thailand provinces, collected from 2012 to 2017. We divided the sample into 2 groups: the first group contains samples from the 3 provinces bordering with 2 countries and the second group from the 2 provinces that do not share a border with any other country. The prevalence of PfKelch13 artemisinin resistance-confirmed mutations, C580Y mutation, mainly found in the provinces located at Thailand–Myanmar border, especially in Ranong, was compared with other sites in this study. The result was consistent with the previous studies [36,38]. Phang Nga, the neighboring province of Ranong, had C580Y mutation in samples collected in 2015. However, the previous study by Putaporntip et al. [39] has reported that Kelch13 mutation in P. falciparum collected in Phang Na at 2009 was wild type. Our result suggested that the spread of artemisinin-resistance in Phang Nga province might be due to the cross-border movements, and mobile populations of people who involved in forest-related activities from Ranong province.
Recently, the study by Kobasa et al. [36] has found that P. falciparum carrying C580Y mutation was present in the samples collected from Chumphon (Thailand–Myanmar border) in 2007. The present study found that P574L mutation, another confirmed mutation for artemisinin resistance [40], was found in Chumphon province in samples from 2014. This mutation was highly prevalent in neighboring countries such as Myanmar and China [41]. In contrast, there was no evidence of PfKelch13 resistance-confirmed mutations in samples from Yala (2017), which is located at the Thailand-Malaysia border. This result was concordant with the previous report in 2015 [40]. In addition, the parasite isolates from Surat Thani, the non-border province also had no mutations in PfKelch13 gene. Our study also indicated that no molecular makers confirmed for artemisinin resistance were detected in Surat Thani and Yala provinces.
Table 2 describes the comparison of different patterns of molecular markers for artemisinin resistance and multidrug resistance between the present study and the other studies. In general, the predominant genotype of P. falciparum isolates in Southern Thailand is the wild type for PfKelch13 gene (85.94%) except for those from Ranong province (11.72%), the province located on Thai-Myanmar border. The recent study indicates the higher percentage of C580Y mutation (38.63%) [36] in the eastern part of Thailand such as Sisaket and Ubon Ratchathani [42] which are close to Cambodia and, Kanchanaburi and Ranong which are at Myanmar border. This implies that the artemisinin-resistant C580Y of P. falciparum was distributed differently between the Myanmar-Thailand and Cambodia-Thailand border regions, with southern Thailand.

Comparison of molecular surveillance on artemisinin resistance and multidrug resistance of Plasmodium falciparum between the present studies and other studies
Since 2015, the first-line treatment for uncomplicated P. falciparum infections in Thailand had been dihydroartemisinin combined with piperaquine (DHA-PQ) [40] which can replace artesunate-mefloquine treatment. However, in vitro and in vivo resistance to mefloquine was highly correlated with the higher copy number of Pfmdr1. The mean copy number of Pfmdr1 in the lower southern region, Yala, was 1.2 while in Ranang, bordering with Myanmar was 2.3 [43]. The distribution of polymorphisms in our study demonstrated that Pfmdr1 isolated from Yala province showed a single pattern N86Y mutation. The samples from Ranong in 2013 and 2014 had the mutations N86Y and Y184F respectively. These results were similar to the results of previous studies by Pickard et al. [23] and Mungthin et al. [43]. Pfmdr1 isolated from Chumphon province in 2014 had only E130K mutation. This mutation was previously recorded in the Cambodia-Vietnam [44] and Thai–Myanmar borders [45]. In Surat Thani province, the mutation of the Pfmdr1 gene was found at N86Y, Y184F, and Y184F plus N1042D positions. Our report would be the first report of mutation profile of Pfmdr1 gene from Surat Thani province. The pattern of Pfmdr1 gene mutations of P. falciparum were similar to the previous study by Mungthin et al. [43], showing that the frequency of N86Y were higher than those of Y184F mutation. In contrast the parasites from east of Thailand carried higher frequency of Y184F [42] (Table 2). However, in the present study, the parasites, isolates of the Pfmdr1 gene were wild type strain distributing in all provinces except for Yala province (N86Y, 100%).
Recently, N86Y and N1042N, SNPs in the Pfmdr1, exhibited a significant reduction of in vitro piperaquine sensitivity while copy number of Pfmdr1 mutations had no effect on piperaquine susceptibility [46]. According to our findings, the pattern of Pfmdr1 isolates from southern Thailand would be mefloquine sensitive but resistant to chloroquine, and those parasites containing the Pfmdr1 N86Y allele would exhibit significant reduction of piperaquine sensitivity. Most parasitic strains isolated from the present study showed the Pfmdr1 N86Y and Y184F mutation from Ranong and Surat Thani were different from those circulated in Ubon Ratchathani, which shares borders with Southern Laos and Northern Cambodia [42] suggesting that the Pfmdr1 mutations isolated from different areas in Thailand carried the different patterns. Hence, both in vivo and in vitro efficacy studies for antimalarial drug monitoring should be done with the samples fromdifferent parts of Thailand.
In summary, the genotyping data in Klech13 gene collected from southern Thailand during 2012–2017 indicated that Ranong province might be responsible for the spread the mutant allele to other areas of the southern Thailand with human movement. This result is supported by the data that approximately half of the malaria cases in Thailand were found in foreign migrant workers, and Ranong province harbors a substantial population of migrant workers in Thailand. This study reported for the first time that C580Y mutation was found in Phang Nga among the samples from 2015. This would be the recent spread of artemisinin resistance allele from Ranong province which has a high frequency of C580Y mutation. The result of SNP patterns in Pfmdr1 is helpful for researchers to monitor changes in parasite genotypes after a change in the treatment regimen. In addition, this data will also provide information for selection of anti-malarial drug policy in Thailand; new candidate drugs should be adopted at least based on the genotype of the parasites in different areas of Thailand.
ACKNOWLEDGMENTS
This work was supported by a grant from Prince of Songkla University, contract no. MET600576S, and the Faculty of Science Research Fund, Prince of Songkla University, contract no. 1-2558-02-008.
Notes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.