Abstract
From October 2015 to August 2018, tapeworm proglottids were obtained from 10 patients who were residents of Daegu and Gyeongbuk provinces and had a history of raw beef consumption. Most of them had no overseas travel experience. The gravid proglottids obtained from the 10 cases had 15–20 lateral uterine branches. A part of internal transcribed spacer 1 (ITS1) DNA of the 10 cases, amplified by polymerase chain reaction (PCR) and digested with AleI restriction enzyme, produced the same band pattern of Taenia saginata, which differentiated from T. asiatica and T. solium. Sequences of ITS1 and cytochrome c oxidase subunit 1 (cox1) showed higher homology to T. saginata than to T. asiatica and T. solium. Collectively, these 10 cases were identified as T. saginata human infections. As taeniasis is one of the important parasitic diseases in humans, it is necessary to maintain hygienic conditions during livestock farming to avoid public health concerns.
-
Key words: Taenia saginata, taeniasis, ITS1, PCR-RFLP, PstI, AleI
INTRODUCTION
Taeniasis is caused by tapeworms belonging to the genus
Taenia and occurs worldwide with a variable degree of prevalence [
1]. Approximately 100 million people are thought to be infected annually with
Taenia [
2]. Humans are generally infected by ingestion of raw or undercooked beef, which contains the larval stage of
T. saginata, and pork may be infected with
T. solium and
T. asiatica. Humans are the only definitive host for these 3 species, whereas cattle and pigs are the intermediate hosts [
2].
The 2 important human pathogens are
T. saginata (beef tapeworm) and
T. solium (pork tapeworm) which have a worldwide distribution, whereas the third species,
T. asiatica (Asian tapeworm) is found only in Asian countries [
3]. The general characteristics of the various
Taenia species are similar, and their eggs are more or less identical. Although there are 4 differentiating morphological features between
T. saginata and
T. asiatica, it is not easy to distinguish between each individual strobila [
4]. Therefore, immunological and/or molecular techniques are used to diagnose taeniasis in humans. Recently, molecular analysis using nuclear ribosomal (e.g., internal transcribed spacer 2 (ITS2), 5.8S rDNA, 28S rDNA, and 18S rDNA) and mitochondrial DNA (e.g., cytochrome c oxidase subunit 1 (
cox1), NADH dehydrogenase subunit 1 (
nad1) cytochrome b (
cytb), and 12S rDNA) were used for the identification of
Taenia tapeworms [
5].
In this paper, we report 10 cases of T. saginata infection from October 2015 to August 2018. The tapeworms were identified by molecular analysis using the PCR-restriction fragment length polymorphism (RFLP) method and partial sequencing of the internal transcribed spacer 1 (ITS1) rDNA region.
CASE RECORD
From October 2015 to August 2018, we covered 10 patients (cases 1–10) had felt the passage of the worms and found whitish yellow tapeworm segments in their underwear or feces. These individuals were admitted to Kyungpook National University Hospital, Keimyung University Dongsan Medical Center, local internal medicine clinic, and Sangju Red Cross Hospital.
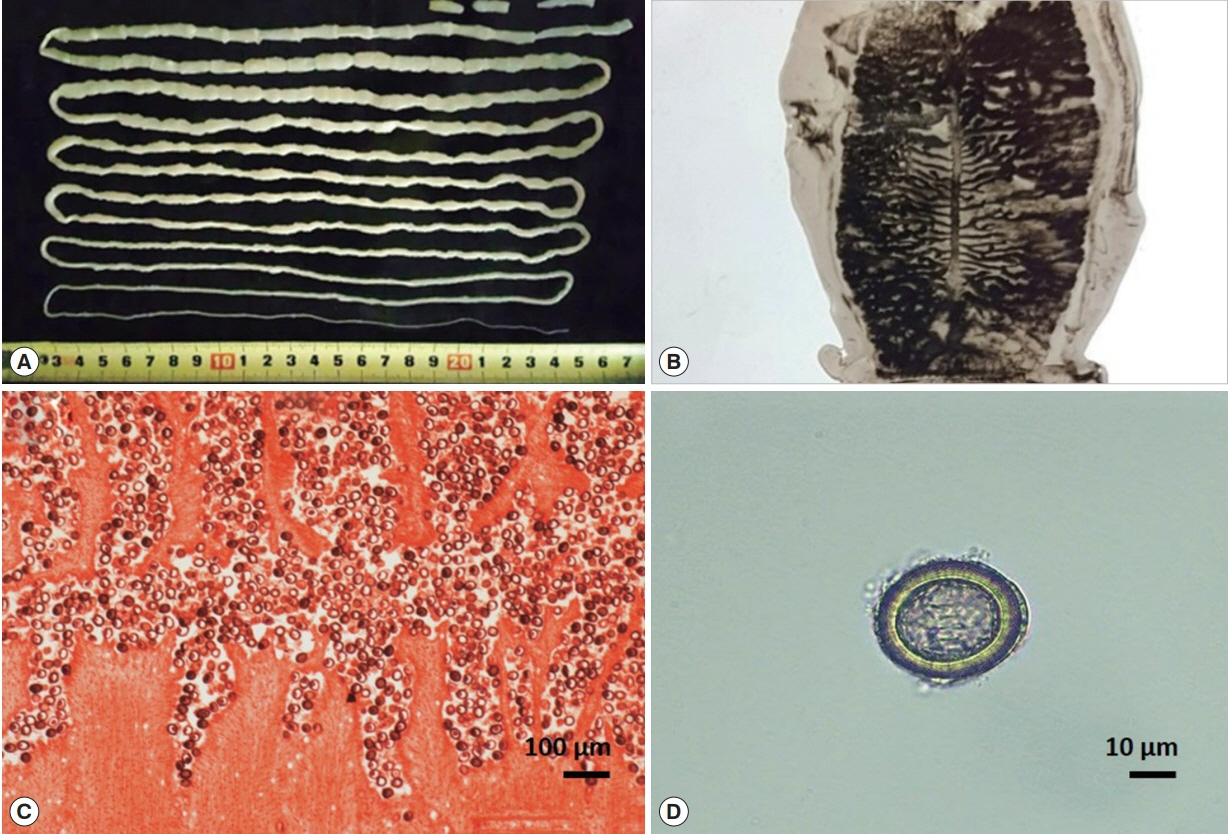
Table 1 summarizes the patient information; almost all the patients had a history of eating raw beef. Most of the patients had no specific clinical symptoms, except for 3 patients who had an itching sensation in their anus. Among them, 8 patients who found worms before getting admitted to the respective hospitals were taking albendazole or mebendazole. However, administration of praziquantel cured them of the parasite. In case 1, whole strobila (approximately 2.7 m in length) without a scolex was obtained after treatment with praziquantel and 40 g magnesium sulfate (MgSO
4) (
Fig. 1A). The gravid proglottids from all the patients, stained with India ink, showed 15–20 lateral uterine branches (
Fig. 1B); and the longitudinal section of gravid proglottids, stained with hematoxylin and eosin, showed that the uterus was filled with eggs (
Fig. 1C, D). However, it was difficult to distinguish between
T. saginata and
T. asiatica by determining the morphological characteristics of their eggs and gravid proglottids. For accurate identification, we developed molecular analyses using a polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) and partial sequencing of ITS1 rDNA region. The genomic DNA was extracted from each tapeworm with a QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Oligonucleotide primers were designed to amplify the ITS1 rDNA (Forward: 5′-CATTGTTGTTGAGCCGAGCCCGG-3′, Reverse: 5′-CAGCAGCAACATCGCCACGT-3′). Size variation was detected by comparing the 512 bp, 513 bp, and 507 bp sequences from
T. saginata, T. asiatica, and
T. solium, respectively. PCR was carried out with PrimeSTAR HS DNA polymerase (Takara, Shiga, Japan) under the following conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 98°C for 10 sec, annealing at 68°C for 5 sec and extension at 72°C for 30 sec, and a final extension at 72°C for 10 min. For RFLP, the purified PCR products were digested with the restriction enzyme
AleI, incubated at 37°C for 2 hr, and then analyzed by electrophoresis on 2% agarose gel.
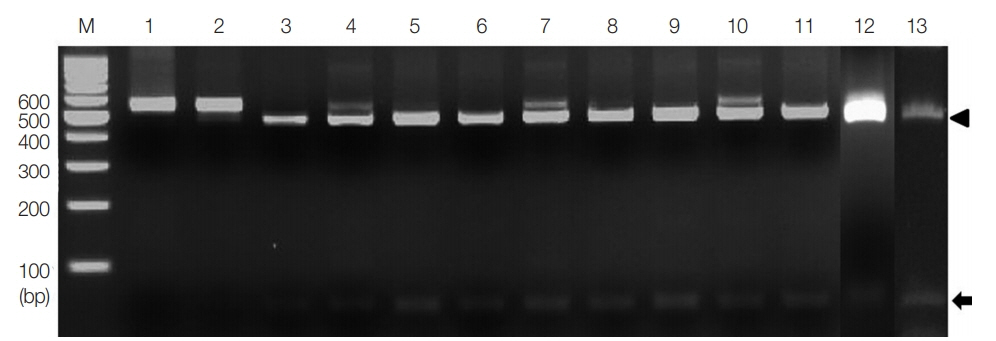
The enzyme
AleI, chosen to clearly distinguish
T. saginata, gave a unique identification pattern for each species. As shown in
Fig. 2, single digestion with
AleI gave 2 different bands, 60 bp and 452 bp (
Fig. 2). These patterns, consistent with
T. saginata (lane 3), were detected in all the 10 specimens (lanes 4–13). However,
AleI could not digest the PCR product of
T. solium and
T. asiatica (lanes 1 and 2).
Additionally, the PCR product was bi-directionally sequenced, and identified using basic local alignment search tool (BLAST) analysis. The ITS1 rDNA sequences of the patients showed 99.4–100% similarity with the partial ITS1 rDNA sequence of
T. saginata (GenBank no. LC004198), 99.0–99.6% with
T. asiatica (GenBank no. KJ207063), and 89.3–89.9% with
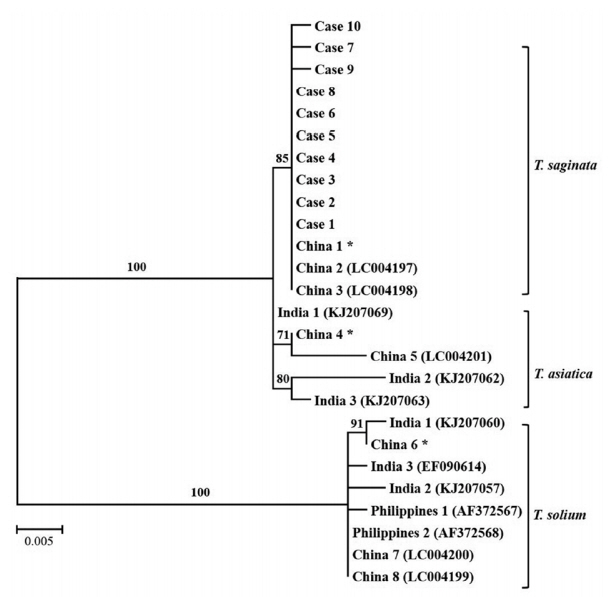
T. solium (GenBank no. AF372567). The phylogenetic relationship was determined using the MEGA 6 program to construct the neighbor-joining tree under the Kimura 2 parameter model. It revealed that all the ITS1 rDNA sequences from this study were clustered within
T. saginata (
Fig. 3).
In order to confirm whether those samples are
T. saginata, cox1 genes were amplified and sequenced for the 10 samples as previously described [
6]. BLAST analysis using the
cox1 sequences of 10 samples showed 99.6–100% homology with previous Korean isolate of
T. saginata (GenBank no. AB465246), but 94.7–95.1% and 88–89% homologies with
T. asiatica (GenBank no. AF445798) and
T. solium (GenBank no. AB494702), respectively. Therefore, molecular diagnosis of all the cases revealed infection with
T. saginata.
DISCUSSION
Taeniasis is diagnosed by recovering the parasite eggs or proglottids in the feces. Although
Taenia spp. eggs are used to identify the family Taeniidae, it cannot be used to distinguish the species [
7]. Identification at the species level is usually based on the number of lateral uterine branches in the gravid proglottids and the presence or absence of hooks on the scolex [
7,
8]. Both
T. saginata and
T. asiatica have 15 to 20 lateral uterine branches, while
T. solium has 7 to 13. No hooks are present on the scolex of
T. saginata, unlike
T. asiatica and
T. solium, which have hooks. Eom and Rim suggested 4 points to distinguish between
T. saginata and
T. asiatica, i.e., the presence or absence of a rostellum on the scolex, the existence of posterior protuberances in gravid proglottids, the number of uterine twigs, and wart-like formations on the bladder wall of the metacestode [
4]. However, the scolex is difficult to obtain due to the worm removal process, and the number of uterine branches in the different species overlaps, making it difficult to distinguish between the species by their morphological characteristics [
8]. Moreover, the examination of the gravid proglottid is not easy without clearing or injection of the uterine branches with India ink.
In order to overcome the limitations in identifying the Taeniid cestodes based on their morphology, several molecular techniques have been developed. Nucleotide sequencing analysis is the most common method and various markers such as nuclear ribosomal RNA genes (18S rRNA [
9], 28S rRNA [
10], and ITS2 [
11]), mitochondrial genes (
cox1 [
10],
cytb [
12],
nad1 [
13], and 12S rRNA [
14]), elongation factor-1-alpha [
15], and ezrin/radixin/moesin-like protein (
elp) genes [
15] were used. Other assays for identifying
Taenia spp. include PCR-RFLP, multiplex PCR [
16], loop-mediated isothermal amplification [
17], random amplified polymorphic DNA analysis [
18], base excision sequence scanning thymine-base analysis [
19], and single-strand conformation polymorphism (SSCP) [
20]. The PCR-RFLP method is preferred as it is a simple, inexpensive, sensitive, and reliable molecular assay compared to nucleotide sequence analysis, which is both time-consuming and costly. In addition, it requires only standard equipment and can be performed quickly [
21]. In several studies, PCR-RFLP using the 5.8S rRNA [
21], ITS2 [
22], 12S rRNA [
23], and
cox1 [
6] gene has been described. In the present study, we used the ITS1 rDNA region to identify the parasites at the species level. Hancock et al. [
24] reported genetic variability in
T. solium by sequencing the COI (cytochrome oxidase I) and ITS1 genes. The COI sequence is conserved in its related country, and ITS1 showed minimal variation within individuals [
24]. Moreover, Dai et al. [
25] demonstrated that the intraspecific sequence variations were 0–0.7% for
cox1 and 0.1–3.6% for ITS among
Spirometra erinaceieuropaei, T. multiceps, and
T. hydatigena isolates from different regions of China, although the interspecific sequence variations were significantly higher (12.1–17.6% and 31–75.7% for
cox1 and ITS, respectively). Therefore, in this study, ITS1 was used for the identification of the 3 species.
The results of PCR-RFLP showed the same digestion pattern in all the specimens and only one nucleotide of ITS1 gene was different in cases 7 and 9 compared to ITS1 sequences of other specimens. Sequence alignment between the ITS1 sequences from the 10 specimens and previously reported ITS1 sequences from the Republic of Korea could not be analyzed as no previous sequence information about ITS1 gene was available. Therefore, as shown in
Fig. 3, sequences of ITS1 gene reported from other countries, such as China, India, and the Philippines were used for the phylogenetic analysis. The phylogenetic analysis showed that the ITS1 genes of all the specimens are clustered in
T. saginata. A similar study had been carried out in Korea using
cox1 gene sequences to compare the patient specimens with the previously reported sequences [
6]. Consistently, the 10 samples in this study also showed 99.6–100% homology with
cox1 sequence of previous Korean isolate of
T. saginata (GenBank no. AB465246).
In Korea, human taeniasis was first reported in 1914; feces examination reported that the prevalence of
Taenia spp. eggs was 7.8%, which remained steady until the 1980s. It gradually decreased and was last reported in 2011 [
26]. However, Cho et al. [
6] reported 4 cases of infection by
T. saginata in 2013 that suggested the reoccurrence of taeniasis in Korea. Similarly, we reported 10 cases of
T. saginata infection over 34 months: 1 case in 2015, 5 cases in 2016, 2 cases in 2017, and 2 cases in 2018, supporting the previous report about the resurgence of
T. saginata infection in Korea. From 1935 to 2005, out of the 68 specimens examined for the identification of the
Taenia species, 51 were identified as
T. asiatica (4 specimens were from Daegu and Gyeongbuk provinces), 14 as
T. saginata, and 3 as
T. solium [
11]. Although
T. asiatica is considered a dominant species, all 10 cases in our study were infected by
T. saginata. All the patients in our study were from Daegu or Gyeongbuk provinces and most of them have no overseas traveling experiences. The 3 patients who had traveled overseas to the Philippines and Hawaii did not eat raw beef there. Therefore, it is necessary to maintain hygiene in the cattle breeding farms and inspect the meat carefully before supplying.
In this study, we developed the PCR-RFLP with the ITS1 rDNA region to identify T. saginata and reported 10 cases of human T. saginata infection in Korea using the developed method. Ribosomal DNA genes, such as ITS1, are useful for identification at the species/strain level. Thus, we hope our study will assist in the diagnosis and identification of the Taenia species, as well as future study related to Taenia infection.
Notes
-
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this study.
Fig. 1Strobila and eggs of Taenia saginata. (A) Whole strobila without the scolex, approximately 2.7 m in length. (B) The gravid proglottid shows the lateral uterine branches (India ink injection). (C) Longitudinal section of a gravid proglottid shows the uterine branches filled with eggs (H & E stain). (D) An egg smeared from a gravid proglottid.
Fig. 2PCR-RFLP of the partial ITS 1 rDNA region digested with restriction enzyme AleI. Lanes 1 and 2, T. solium and T. asiatica; lanes 4–13, cases 1–10. Digestion of the PCR products with AleI produced 60 bp (arrow) and 452 bp (arrowhead) fragments compatible to T. saginata (lane 3). M, 100 bp DNA marker.
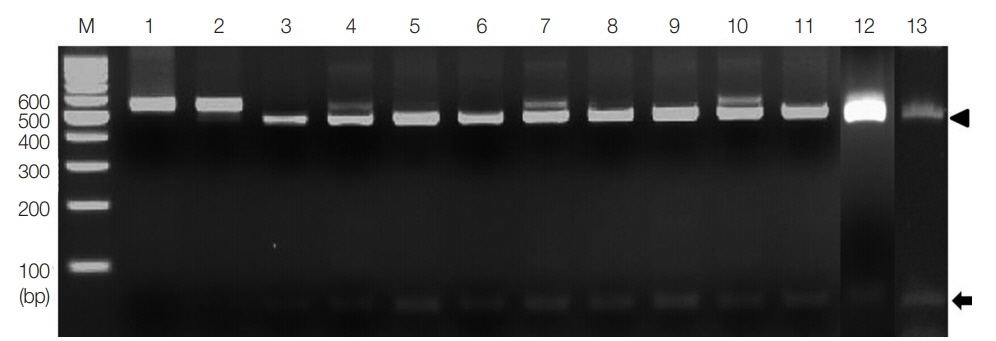
Fig. 3A phylogenetic tree of human Taenia cases based on ITS 1 rDNA sequences. Numbers on the branches are the bootstrap values for 1,000 replications. *China 1, 4, and 6 are ITS 1 sequences of T. saginata, T. asiatica, and T. solium from the Parasite Resource Bank, Chungbuk National University.
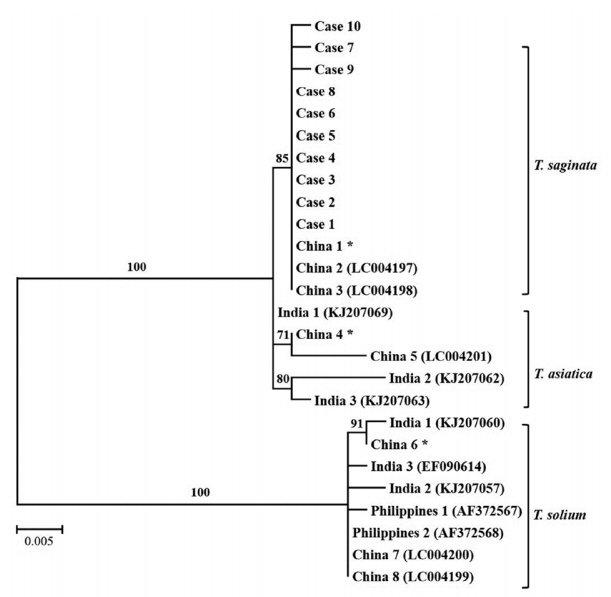
Table 1Summary of ten cases of Taenia saginata infection
Table 1
|
Case No. |
Age (y)/Sex |
Date of first symptom appeared |
Visited date on the hospital |
Main complaint |
Suspected source of infection |
Visited country |
|
1 |
57/F |
- |
Oct-15 |
Perianal itching |
Raw beef |
- |
|
2 |
70/F |
Aug-15 |
Apr-16 |
- |
Raw beef |
- |
|
3 |
29/F |
Sep-16 |
Oct-16 |
- |
*
|
- |
|
4 |
45/F |
Sep-16 |
Oct-16 |
Perianal itching |
Raw beef |
The Philippines |
|
5 |
33/M |
- |
Nov-16 |
- |
Raw beef |
- |
|
6 |
26/M |
Oct-16 |
Nov-16 |
- |
Raw beef |
The Philippines |
|
7 |
23/M |
Aug-16 |
Feb-17 |
- |
Raw beef |
- |
|
8 |
74/F |
Jan-17 |
Mar-17 |
Perianal itching |
Raw beef |
- |
|
9 |
27/F |
-2016 |
Jan-18 |
- |
*
|
Hawaii |
|
10 |
25/F |
Aug-17 |
Aug-18 |
- |
*
|
- |
References
- 1. Macpherson CNL, Craig PS. The zoonotic Taeniae of Africa. Parasitic Helminthes and Zoonoses in Africa. Dordrecht, Netherlands. Springer; 1991, pp 54-82.
- 2. Melkie G, Tewodros AE. Review on Taeniasis and its zoonotic importance. Europ J Appl Sci 2015;7:182-191.
- 3. Galán-Puchades MT, Fuentes MV.
Taenia asiatica: the most neglected Human Taenia and the possibility of cysticercosis. Korean J Parasitol 2013;51:51-54.
- 4. Eom KS, Rim HJ. Morphologic descriptions of Taenia asiatica sp. n. Korean J Parasitol 1993;31:1-6.
- 5. Ale A, Victor B, Praet N, Gabriël S, Speybroeck N, Dorny P, Devleesschauwer B. Epidemiology and genetic diversity of Taenia asiatica: a systematic review. Parasit Vectors 2014;7:45.
- 6. Cho J, Jung BK, Lim H, Kim MJ, Yooyen T, Lee D, Eom KS, Shin EH, Chai JY. Four cases of Taenia saginata infection with an analysis of cox1 gene. Korean J Parasitol 2014;52:79-83.
- 7. The Center for Food Security and Public Health. Taenia Infections [Internet]; Available from: http://www.cfsph.iastate.edu/Factsheets/pdfs/taenia.pdf
- 8. Eom KS. What is Asian Taenia? Parasitol Int 2006;55(suppl):137-141.
- 9. Yan H, Lou Z, Li L, Ni X, Guo A, Li H, Zheng Y, Dyachenko V, Jia W. The nuclear 18S ribosomal RNA gene as a source of phylogenetic information in the genus Taenia
. Parasitol Res 2013;112:1343-1347.
- 10. Bowles J, McManus DP. Genetic characterization of the Asian Taenia, a newly described taeniid cestode of humans. Am J Trop Med Hyg 1994;50:33-44.
- 11. Jeon HK, Kim KH, Chai JY, Yang HJ, Rim HJ, Eom KS. Sympatric distribution of three human Taenia tapeworms collected between 1935 and 2005 in Korea. Korean J Parasitol 2008;46:235-241.
- 12. Yamasaki H, Nakao M, Sako Y, Nakaya K, Ito A. Molecular identification of Taenia solium cysticercus genotype in the histopathological specimens. Southeast Asian J Trop Med Public Health 2005;36:131-134.
- 13. Gasser RB, Zhu X, McManus DP. NADH dehydrogenase subunit 1 and cytochrome c oxidase subunit I sequences compared for members of the genus Taenia (Cestoda). Int J Parasitol 1999;29:1965-1970.
- 14. Roelfsema JH, Nozari N, Pinelli E, Kortbeek LM. Novel PCRs for differential diagnosis of cestodes. Exp Parasitol 2016;161:20-26.
- 15. Okamoto M, Nakao M, Blair D, Anantaphruti MT, Waikagul J, Ito A. Evidence of hybridization between Taenia saginata and Taenia asiatica
. Parasitol Int 2010;59:70-74.
- 16. Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya K, Qiu D, Mamuti W, Craig PS, Ito A. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J Clin Microbiol 2004;42:548-553.
- 17. Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J Clin Microbiol 2009;47:168-174.
- 18. Eom KS, Jeon HK, Kong Y, Hwang UW, Yang Y, Li X, Xu L, Feng Z, Pawlowski ZS, Rim HJ. Identification of Taenia asiatica in China: molecular, morphological, and epidemiological analysis of a Luzhai isolate. J Parasitol 2002;88:758-764.
- 19. Yamasaki H, Nakao M, Sako Y, Nakaya K, Sato MO, Mamuti W, Okamoto M, Ito A. DNA differential diagnosis of human taeniid cestodes by base excision sequence scanning thymine-base reader analysis with mitochondrial genes. J Clin Microbiol 2002;40:3818-3821.
- 20. Gasser RB, Zhu X, Woods W. Genotyping Taenia tapeworms by single-strand conformation polymorphism of mitochondrial DNA. Electrophoresis 1999;20:2834-2837.
- 21. Mayta H, Talley A, Gilman RH, Jimenez J, Verastegui M, Ruiz M, Garcia HH, Gonzalez AE. Differentiating Taenia solium and Taenia saginata infections by simple hematoxylin-eosin staining and PCR-restriction enzyme analysis. J Clin Microbiol 2000;38:133-137.
- 22. Gasser RB, Chilton NB. Characterisation of taeniid cestode species by PCR-RFLP of ITS2 ribosomal DNA. Acta Trop 1995;59:31-40.
- 23. Rodriguez-Hidalgo R, Geysen D, Benítez-Ortiz W, Geerts S, Brandt J. Comparison of conventional techniques to differentiate between Taenia solium and Taenia saginata and an improved polymerase chain reaction-restriction fragment length polymorphism assay using a mitochondrial 12S rDNA fragment. J Parasitol 2002;88:1007-1011.
- 24. Hancock K, Broughel DE, Moura IN, Khan A, Pieniazek NJ, Gonzalez AE, Garcia HH, Gilman RH, Tsang VC. Sequence variation in the cytochrome oxidase I, internal transcribed spacer 1, and Ts14 diagnostic antigen sequences of Taenia solium isolates from South and Central America, India, and Asia. Int J Parasitol 2001;31:1601-1607.
- 25. Dai RS, Liu GH, Song HQ, Lin RQ, Yuan ZG, Li MW, Huang SY, Liu W, Zhu XQ. Sequence variability in two mitochondrial DNA regions and internal transcribed spacer among three cestodes infecting animals and humans from China. J Helminthol 2012;86:245-251.
- 26. Chai JY. Human taeniasis in the Republic of Korea: hidden or gone? Korean J Parasitol 2013;51:9-17.