Mitochondrial DNA Sequence Variability of Spirometra Species in Asian Countries
Article information
Abstract
Mitochondrial DNA sequence variability of Spirometra erinaceieuropaei in GenBank was observed by reinvestigation of mitochondrial cox1 and cytb sequences. The DNA sequences were analyzed in this study, comprising complete DNA sequences of cox1 (n=239) and cytb (n=213) genes. The 10 complete mitochondrial DNA sequences of Spirometra species were compared with those of Korea, China and Japan. The sequences were analyzed for nucleotide composition, conserved sites, variable sites, singleton sites and parsimony-informative sites. Phylogenetic analyses was done using neighbor joining, maximum parsimony, Bayesian inference and maximum-likelihood on cox1 and cytb sequences of Spirometra species. These polymorphic sites identified 148 (cox1) and 83 (cytb) haplotypes within 239 and 213 isolates from 3 Asian countries. Phylogenetic tree topologies were presented high-level confidence values for the 2 major branches of 2 Spirometra species containing S. erinaceieuropaei and S. decipiens, and S. decipiens sub-clades including all sequences registered as S. erinaceieuropaei in cox1 and cytb genes. These results indicated that mitochondrial haplotypes of S. erinaceieuropaei and S. decipiens were found in the 3 Asian countries.
INTRODUCTION
The tapeworm Spirometra erinaceieuropaei is the most well-known species of the genus Spirometra tapeworms. Its plerocercoid larvae can infect the brain, eyes, breast, spinal cord, and subcutaneous tissue of humans [1,2]. Human sparganosis is a worldwide parasitic zoonosis, which causes serious clinical diseases. These diseases have been reported in more than 1,600 cases, mostly in east and Southeast Asia, and sporadic cases in South America, Europe and Africa [3]. Precise identification of these spirometrid tapeworms is therefore important in efforts to control the diseases caused by human sparganosis.
Species classification of the tapeworms in the genus Spirometra remains controversial. Identification of spirometrid species within the genus Spirometra species has been attempted by many researchers. Six Spirometra species were initially studied and morphologically identified [4]. The new species S. mansonoides derived from cat in the Syracuse region of the USA was then reported [5]. Fourteen Spirometra species were recognized by 2 separate groups as bresslauei and okumurai [6]. A genotype of all species in the genus Spirometra was the same as S. erinaceieuropaei [7]. The morphological difference in the uterine shape of the mature proglottids of S. erinaceieuropaei is due to differences in developmental larval stages [8]. S. erinaceieuropaei, S. pretoriensis, S. theileri, and S. mansonoides were the only valid species [9]. More recently, human sparganosis was identified by morphological and genetic analyses, and the complete mitochondrial genomes of S. erinaceieuropaei and S. decipiens have been recorded and compared in Korea [10,11].
Molecular identification has played an important role in studies on phylogenetics, parasite genetic variation and evolution over the decades. Mitochondrial DNA sequences have provided genetic markers for phylogenetic reconstruction, taxonomic identification, population genetics, and epidemiological investigations [12]. Investigations of phylogenetic relationships and genetic variation of the Spirometra tapeworms employed mitochondrial genomes or genes such as cytochrome c oxidase subunit 1 and 3 (cox1 and cox3) and NADH dehydrogenase subunit 1, 3, and 4 (nad1, nad3, and nad4) [2,10,11,14–20].
The nucleotide sequence data employed by the aforementioned studies were obtained from public databases. The availability of nucleotide sequences of Spirometra in public databases has expanded since the 1990s and now includes the DNA DataBank of Japan (DDJB), European Molecular Biology Laboratory (EMBL) and GenBank (National Center for Biotechnology Information). These organizations provide a comprehensive public database of nucleotide sequences and supporting bibliographic and biological annotation of the submitted DNA sequence data from authors and from the bulk submission of expressed sequence tag (EST), genome survey sequence (GSS), whole-genome shotgun (WGS), and other high-throughput data from sequencing centers [21]. The sequence data in these public databases is made available by persons or groups that continue to make corrections to or update the data. Therefore, the speciation of sister or congeneric species may need to be reconsidered and revised based on any morphological analyses.
A total of 483,611 nucleotide sequences of S. erinaceieuropaei have been deposited in the GenBank database. Of those, 908 protein-coding DNA sequences of S. erinaceieuropaei are found in GenBank including nuclear and mitochondrial genomic DNA, as of November 2018. As indicated above, these DNA sequences may need to be revised following careful reconsideration based on morphological analyses of Spirometra tapeworms. The aim of this study was to observe mitochondrial DNA sequence variability of S. erinaceieuropaei in GenBank by reinvestigation of the sequence analysis of mitochondrial DNA based on species identified by morphological analyses of S. erinaceieuropaei and S. decipiens.
MATERIALS AND METHODS
DNA sequences of Spirometra spp. in GenBank
A total of 1,024 mitochondrial DNA sequences were extracted from GenBank in November 2018 for mitochondrial genes, cox1 (n=514), cox2 (n=10), cox3 (n=73), cytb (n=213), nad1 (n=60), nad2 (n=10), nad3 (n=10), nad4 (n=74), nad4L (n=10), nad5 (n=30), nad6 (n=10), and atp6 (n=10). Nineteen cox1 sequences were obtained of human sparganosis from Australia, Japan, Korea and Thailand. The remaining cox1, cox3, cytb, nad1, and nad4 sequences were obtained from various animals such as fox, dog, cat, snake, and frog. Four hundred and fifty-two DNA sequences were analyzed in this study, comprising DNA sequences of the cox1 gene from Australia (n=12), China (n=360), India (n=1), Indonesia (n=3), Japan (n=30), Korea (n=3), Laos (n=16), New Zealand (n=1), Myanmar (n=2) and Thailand (n=14), and 213 DNA sequences of the cytb gene from China (n=209), Japan (n=2) and Korea (n=2). Among them, a total of 239 (cox1) and 213 (cytb) complete DNA sequences were inferred phylogenetic analysis (Table 1). The 10 complete mitochondrial DNA sequences of Spirometra species were compared with those of KJ599680 (S. erinaceieuropaei) and KJ599679 (S. decipiens), and comprised sequences from China (JQ267473, KY114886, KY114887, KY114888, KY114889 and KU852381) and Japan (AB374543 and AP017668), which were used as reference criteria for genetic observation of the GenBank database.
MtDNA sequence analyses
DNA sequences were assembled using the Geneious 9.0 software program (Biomatter, Auckland, New Zealand) and than aligned using MAFFT methods in the Geneious 9.0 software program by comparison with DNA sequences of S. erinaceieuropaei and S. decipiens in the GenBank database. Molecular Evolution Genetics Analysis (MEGA) software version 7.0 was employed to analyze nucleotide composition, conserved sites, variable sites, singleton sites and parsimony-informative sites [22]. Evaluation of the number of haplotypes, nucleotide diversity and haplotype diversity were performed using DnaSP software version 6.12 [23].
Phylogenetic analyses
Phylogenetic analysis was evaluated using neighbor joining (NJ), maximum parsimony, Bayesian inference (BI) and maximum-likelihood (ML) using cox1 (1,566 bp) and cytb (1,110 bp) sequences of Spirometra species. NL analysis was performed using MEGA version 7.0 [20]. MP analysis was performed in PAUP4b10 [24] using heuristic searches with tree bisection reconnection (TBR) branch swapping. ML analyses of cox1 and cytb used RAxML version 7.3.1 [25] after the TRN+G substitution model was chosen with Modeltest using Partition Finder version 2.1.1 [26]. BI analyses were used in MrBayes 3.2 [27] after the HKY+G substitution model was chosen with Modeltest using Partition Finder version 2.1.1 and then running 2 independent MC3 runs of 4 Markov chains each, for 10 million metropolis-coupled Markov chain Monte Carlo (MCMC) generations and discarding the first 25% generation as burn-in every 1,000 generations. Phylogenetic trees were constructed using the mitochondrial cox1 and cytb DNA sequences of Spirometra species in GenBank and 3 taxa of Diphyllobothriidae represented by Dibothriocephalus latum (NC_008945) and D. nihonkaiense (NC_009463) as the outgroup to root the resulting trees.
RESULTS
Divergences of mt genomes among Spirometra
Mitochondrial genomes of Spirometra tapeworms have been reported for S. erinaceieuropaei 13,643 bp (AB374543; Japan), 13,603 bp (AP017668; Japan), 13,631 bp (JQ267473; China), 13,643 bp (KJ599680; Korea), 13,569 bp (KU852381; China), 13,609 bp (KY114887; China), 13,680 bp (KY114888; China), 13,643 bp (KY114886; China), 13,680 bp (KY113889; China), and for S. decipiens 13,641 bp (KJ599679, Korea). The mt genomes each contained 36 genes comprising 12 protein-coding genes, 22 tRNAs and 2 rRNAs.
A percentage pairwise comparison of sequence divergences of the 12 protein-coding genes between S. erinaceieuropaei (KJ 599680) and S. decipiens (the remaining mt genomes in this study) is shown in Table 2. The 12 protein-coding genes comprised 10,065 bp and 3,355 codons (S. erinaceieuropaei) and 10,067 bp and 3,355 codons (S. decipiens) of their respective mitochondrial genomes. The overall nucleotide sequence divergence of the 12 protein-coding genes between S. erinaceieuropaei and S. decipiens differed by 13.0% (Table 2). The divergence of amino acid sequences of the 12 protein-coding genes of S. erinaceieuropaei and S. decipiens ranged from as low as 2.3% (nad4L) to as high as 14.8% (nad6). The nucleotide sequence divergence of the most variable gene was the nad6 and the most highly conserved gene was cox1, which was 18.8% and 9.4%, respectively, when comprising S. erinaceieuropaei and S. decipiens (Table 3). The nucleotide sequence difference of the 12 protein-coding genes within interspecies of S. decipiens between Sd and Sd* in Table 3 was 1.5% and ranged from as low as 0.0% (cox2 and nad6) to as high as 2.6% (nad4) (Table 3).

Percentage whole mitochondrial DNA sequence homology of Spirometra species from GenBank as of November 2018
Sequence analysis
The concatenated sequence analysis contained 239 (cox1) and 213 (cytb) sequences, of which 1,566 bp and 1,110 bp, respectively, comprised mitochondrial genes. Within the cox1 and cytb genes was found 198 and 93 polymorphic sites, 166 and 69 parsimony informative sites, and 32 and 24 singleton variable sites, respectively. The polymorphic sites identified comprised 148 (cox1) and 83 (cytb) haplotypes within 239 and 213 isolates, respectively, from 3 Asian countries (China, Japan and Korea). Non-synonymous substitution sites found were 1,187.98 (cox1) and 836.36 (cytb), while 363.02 and 270.64 represented synonymous substitution sites. The genetic divergence of cox1 and cytb sequences of Spirometra species (S. decipiens) ranged from 0% to 5.9% and 0% to 3.6%, respectively. These 2 mitochondrial genes have high Hd (haplotype diversity) and Pi (nucleotide diversity) (Table 4).
Phylogenetic diversity
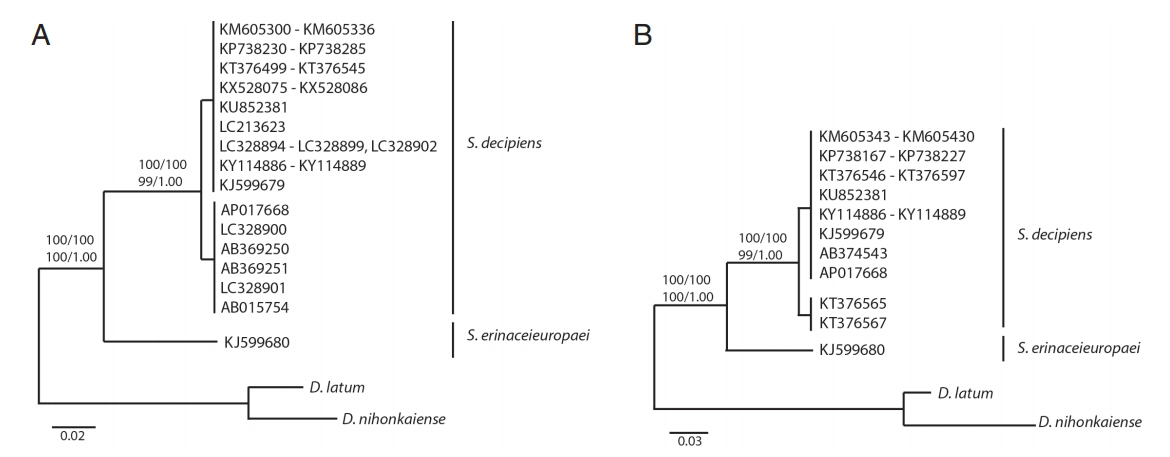
All haplotypes of Spirometra species were separated into 2 distinct clades in phylogenetic analyses based on neighbor joining, maximum parsimony, maximum likelihood and Bayesian inference. Clade I was S. erinaceieuropaei and clade II was S. decipiens. Phylogenetic tree topologies generated from the 4 analytic methods were identical and presented high-level confidence values for the 2 major branches of 2 Spirometra species for the cox1 and cytb genes. The ML and BI analyses supported monophyly of Spirometra species and identified the species S. erinaceieuropaei and S. decipiens as 2 clades with bootstrap values of 100 and 1.00, respectively, based on cox1 and cytb sequences (Fig. 1). The tree topologies of the NJ and MP analysis were congruent with those topologies in the ML and BI analyses. The bootstrap value of NJ and MP was 100 and 100, respectively. Both of tree in placing the 2 sub-clade were observed in S. decipiens clade by ranged 0–5.9% (cox1) and 0–3% (cytb) DNA sequences differences (Fig. 1).
DISCUSSION
Morphological and experimental studies for Spirometra species have been reported by Chandler [28] and Faust et al. (1929) [4]. Faust et al. (1929) reviewed Spirometra species (under the name Diphyllobothrium) with morphological and biological studies of S. erinaceieuropaei (Rudolphi, 1819), S. decipiens (Diesing, 1850), S. ranarum (Gastaldi, 1854), S. mansoni (Cobbold, 1882), S. houghtoni (=S. mansoni, Faust et al. [4], 1929) and S. okumurai (Faust et al. [4], 1929). S. erinaceieuropaei was first reported by Rudolphi (1819) and assigned the name Dubium erinacei-europaei from thoracic spargana of the hedgehog (Erinaceus europaeus). This larval tapeworm was recognized as a sparganum by Diesing (1854) and renamed to Sparganum lanceolatum by Moiln (1859). S. decipiens was described by Diesing (1854) and Lühe (1899) reexamined the original material with the recorded uterine character which facilitated its identification. Detailed morphological characteristics were then described by Chandler (1925) [4,28]. The only Spirometra species reported in North America was by Mueller (1935), who described the new species S. mansonoides, which was distinguished from S. mansoni distributed in the Asian region based on morphological characteristics [29].
The major difference in feature between S. erinaceieuropaei and S. decipiens is the spirally coiled uterus of S. erinaceieuropaei, which consists of 5–7 coils, while that of S. decipiens consist of 4–4.5 coils [4,10]. However, certain morphological characteristics are by S. decipiens and S. mansoni, such as their male and female reproductive organs. Morphological characteristics of S. ranarum include a uterine morphology that consists of 3–4.5 uterine coils, of which the posterior 2 are broader and more dilated than the terminal uterine ball [4,30]. One notable feature of S. mansonoides is the C-shaped outer loop of the uterus with its anterior limb constricted in the midline to form a lateral expulsion chamber [29].
Identification of Spirometra species has been reported from many endemic areas in Asian, South American and African countries as a result of molecular studies based on mitochondrial DNA sequences. Partial mitochondrial DNA sequence variations of S. erinaceieuropaei isolates from China, Japan and Indonesia ranged from 0.0–2.6% [31], 0.0–3.1% [14,15], 0.0–8.4% [16], and 0.0–7.4% [20] for cox1, 0.0–1.5% [15], 0.0–2.4% [16,20] for cox3, 0.0–2.8% [15] for nad1, and 0.0–2.7% [15], 0.0–1.4% [20] for nad4. Complete mitochondrial DNA sequence variations of S. erinaceieuropaei ranged from 0% to 4.9% for cox1 and 0% to 3.6% for cytb [17,18]. African human sparganosis was found to differ from that of Asian and South American isolates by analysis of mitochondrial DNA sequence data [32]. Two Spirometra species, S. theileri (Baer, 1924) and S. pretoriensis (Baer, 1924) were reported in lions and hyenas in Africa [33]. South American isolates of Spirometra were neither S. erinaceieuropaei, S. decipiens nor S. mansonoides by cox1 sequence analysis. Five Spirometra species comprising S. decipiens, S. mansoni, S. gracile (Baer, 1927), S. longicollis (Parodi and Widakowich, 1917) and S. mansonoides were reported from wild animals in South America [34]. These studies provoked epidemiological questions concerning the need to identify which Spirometra species are distributed in endemic areas and which Spirometra species induce human sparganosis. Many previous studies may need to be reexamined using a combination of molecular and morphological techniques to better understand the epidemiological status of Spirometra spp.
In this study, complete mitochondrial DNA sequence analyses were performed using cox1 and cytb genes and mitochondrial genomes derived from the GenBank database, thereby allowing for genetic variation and phylogenetic relationships to be determined. The nucleotide diversity (Pi) value is employed as a measure of genetic variation, and Pi>0.01 is considered to indicate a comparatively large variation in animal phyla. The Pi values (genetic diversity) of the cox1 and cytb genes were greater than 0.01 in this study, suggesting that nucleotide sequences of Spirometra species in the GenBank database have a large variation. The results of genetic divergence and polymorphic sites showed that there is moderate to high genetic differentiation. Sequence differences of Spirometra sp. haplotypes was shown to be greater than 5% for data in the GenBank database. These results indicated that mitochondrial haplotypes of Spirometra species were represented by at least 2 haplotypes within S. erinaceieuropaei. The genetic distance between S. erinaceieuropaei and S. decipiens differed by 12.4% in the 12 protein-coding genes, whereas the sequence difference for the complete mitochondrial sequences was 12.9% [35]. The phylogenetic analyses based on the cox1 and cytb genes, and analyses of complete mt genome sequences based on ML, BI, NJ and MP methods, all suggested that mitochondrial DNA sequences of Spirometra species were separated into 2 distinct haplotypes isolated from China, Japan and Korea. Phylogenetic tree topologies showed 2 major branches of 2 Spirometra species containing S. erinaceieuropaei and S. decipiens, and S. decipiens sub-clades including all sequences registered as S. erinaceieuropaei in the cox1 and cytb genes from GenBank.
This study has shown that the overall nucleotide sequence and amino acid divergence of 12 protein-coding genes between S. erinaceieuropaei and S. decipiens differed by 12.9% and 12.4%, respectively. Analyses of the cox1 and cytb genes revealed 198 and 93 polymorphic sites, of which 166 and 69 were parsimony informative sites, and 32 and 24 were singleton variable sites, respectively. These polymorphic sites showed 148 (cox1) and 83 (cytb) haplotypes within 239 and 213 isolates from 3 Asian countries (China, Japan and Korea). Phylogenetic tree topologies generated from the 4 analytic methods were identical and presented high-level confidence values for the 2 major branches of 2 Spirometra species containing S. erinaceieuropaei and S. decipiens, and S. decipiens sub-clades including all sequences registered as S. erinaceieuropaei for the cox1 and cytb genes from GenBank. These results indicated that mitochondria haplotypes of S. erinaceieuropaei and S. decipiens were found in Asian countries.
ACKNOWLEDGMENT
This work was supported by the National Research Foundation of Korea (NRF-2017K1A3A1A09085607).
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.