Proteomic Analysis on Exosomes Derived from Patients’ Sera Infected with Echinococcus granulosus
Article information
Abstract
Cystic echinococcosis (CE), a zoonotic disease caused by Echinococcus granulosus at the larval stage, predominantly develops in the liver and lungs of intermediate hosts and eventually results in organ malfunction or even death. The interaction between E. granulosus and human body is incompletely understood. Exosomes are nanosized particles ubiquitously present in human body fluids. Exosomes carry biomolecules that facilitate communication between cells. To the best of our knowledge, the role of exosomes in patients with CE is not reported. Here, we isolated exosomes from the sera of patients with CE (CE-exo) and healthy donors and subjected them to liquid chromatography-tandem mass spectrometry analysis. Proteomic analysis identified 49 proteins specifically expressed in CE-exo, including 4 proteins of parasitic origin. The most valuable parasitic proteins included tubulin alpha-1C chain and histone H4. And 8 proteins were differentially regulated in CE-exo (fold change>1.5), as analyzed with bioinformatic methods such as annotation and functional enrichment analyses. These findings may improve our understanding about the interaction between E. granulosus and human body, and may contribute to the diagnosis and prevention of CE.
INTRODUCTION
Cystic echinococcosis (CE), a zoonotic disease caused by Echinococcus granulosus at the larval stage, causes significant economic losses to the cattle industry and has a great impact on human health [1,2]. Human infections are associated with the oral intake of parasite eggs. The eggs hatch in the digestive tract, penetrate the intestine walls, enter the blood circulation, and eventually settle mostly in the liver and lungs. This disease has been recognized for more than 2,000 years, and is endemic in cattle-producing areas, including China. E. granulosus is typically transmitted between dogs and its second host, human beings. The detection of infection is difficult in the beginning, as the growth of hydatid cysts is slow with almost no symptom for many years [3]. In the late stage of infection, a fluid-filled bladder-like structure with inner germinal layer is detected in the abdomen of the patient, mostly in the liver. The complete excision of the fluid-filled bladder-like structure is often difficult and the risk of recurrence is high [4]. The lack of early diagnosis and effective treatment regimens for CE may result in organ malfunction and sometimes even death. Therefore, the diagnosis of CE is important to improve the prognosis of patients with CE. During infection, Echinococcus has the ability to evade the immune system and successfully reside in the human body; the mechanism involved in the parasite establishment, growth, and persistence is still largely unknown [5,6].
Exosomes are 30–150-nm membranous vesicles released by most cell types, and could be detected in all body fluids, including urine, blood, milk, saliva, amniotic fluid, sperm, and follicular fluid [7]. Exosome biogenesis involves the inward budding of multivesicular bodies. After fusion with the plasma membrane, the multivesicular bodies release exosomes into the extracellular environment. Exosomes contain a variety of molecules such as proteins, nucleic acids, and lipids [8], all of which reflect their origin. The cargo molecules vary depending upon the parent cells, and the selectively packaged functional biomolecules, including enzymes, cytoskeletal proteins, and lipids, could alter the recipient cell [9,10]. Exosomes were originally considered as containers to remove unwanted molecules from their parent cells, and subsequent studies demonstrated their antigen-presenting ability [11]. These vesicles are thought to play crucial roles in intercellular communication between cells and in many pathological conditions such as various types of cancers and autoimmune diseases [12]. Exosomes are potentially useful for the diagnosis of diseases such as cancer [13–15]. The release of exosomes has been demonstrated in many parasitic infections. Protein analysis of the exosomes derived from Plasmodium falciparum has led to the identification of biomarkers of infection [16]. Toxoplasma gondii exosomes could modulate the activation of macrophages and trigger cellular and humoral immune responses associated with protection against parasite infection [17,18]. Schistosome-derived exosomes play an important role in host-parasite interaction and may serve as useful tools in the development of therapeutics and vaccines [19]. The characteristics of exosomes isolated from hydatid fluid of sheep have been recently elaborated [20]. The research on the exosomes derived from patients with CE is of clinical significance. In this study, we performed proteomic analysis of the exosomes isolated from patients with CE and healthy donors for the better understanding of CE pathogenesis.
MATERIALS AND METHODS
Blood collection
Two to five ml of peripheral blood were collected from 3 patients with CE at the General Hospital of Ningxia Medical University, with informed consent (Table 1). Peripheral blood samples were also collected from healthy donors.
Isolation and purification of exosomes
Exosomes were isolated and purified from serum using ExoQuick kit (SBI, California, USA), a proven alternative to ultracentrifugation [21–23], according to the manufacturer’s instructions. Briefly, serum samples were centrifuged at 3,000×g for 15 min to remove cells and debris. Add appropriate volume of ExoQuick exosome precipitation to supernatant of serum and mix well by inverting. Refrigerate 30 min at 4°C, and centrifuge mixture at 1,500×g for 30 min. After centrifugation, the exosomes may appear as a beige or white pellet at the bottom of vessel. Spin down residual ExoQuick solution by centrifugation at 1,500×g and aspirate supernatant. Resuspended exosome pellet in sterile 1×phosphate-buffered saline (PBS) and either stored at −80°C or directly used.
Transmission electron microscopy (TEM)
Exosome samples were subjected to phosphotungstic acid negative staining. About 25 μl of exosome sample was dropped on a carbon-coated grid. After 2 min, the grid was incubated with 2% phosphotungstic acid for 2 min, and its edges were dried with a filter paper. The exosomes were visualized with TEM (HITACHI-H7650, Tokyo, Japan).
Nanoparticle tracking analysis (NTA)
The exosome size distribution analysis and quantification were performed with a zeta view instrument (Particle Metrix) and the corresponding software.
Western blot analysis
In total, 20 μg of exosomes was separated by 12% SDS-PAGE, and the protein bands were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, Massachusetts, USA). After 2 hr of incubation in 5% skim milk in PBST (0.1% Tween 20 in PBS, PH 7.3), the membranes were incubated overnight at 4°C with primary antibodies: anti-CD9 (1/1,000) or anti-CD63 (1/300). Primary antibodies against CD63 and CD9 were purchased from Abcam (Cambridge, Massachusetts, USA). Anti-GAPDH were obtained from BIOSS (Beijing, China). The membrane was subsequently incubated with a horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Abcam) as secondary antibody (1/3,000). The membranes were developed using an enhanced chemiluminescence western blot detection system. The electrophoresis of unstained gel was performed using TGX Stain-Free™ FastCast™ Acrylamide Kit (Bio-rad, Hercules, California, USA).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and loaded onto a home-made reversed-phase analytical column. The gradient included an increase in solvent B (0.1% formic acid in 98% acetonitrile) from 6% to 23% over 26 min, 23% to 35% in 8 min, and then to 80% in 3 min, followed by a hold at 80% for the last 3 min at a constant flow rate of 400 nl/min on EASY-NLC 1000 UPLC system.
The peptides were subjected to nanospray ionization (NSI) source followed by tandem mass spectrometry (MS/MS) on Q Exactive™ Plus (Thermo, Massachusetts, USA) coupled online to an ultra-performance liquid chromatography (UPLC) system. The electrospray voltage applied was 2.0 kV and the m/z scan range were 350 to 1,800 for full scan. Intact peptides were detected in the orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using normalized collision energy (NCE) setting as 28, and the fragments were detected in the orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 sec dynamic exclusion was employed. Automatic gain control was set at 5E4.
Gene ontology (GO) annotation
GO annotation was performed using the UniProt-GOA database (www.http://www.ebi.ac.uk/GOA/). The intensified protein IDs were converted to UniProt ID and subjected to GO mapping. The identified proteins that were not annotated with UniProtGOA, were subjected to InterProScan based on the GO function using the protein sequence alignment method. The proteins were classified with GO annotation into 3 categories, namely, biological process, cellular component, and molecular function.
Domain annotation
The functional description of the identified protein domain was annotated with InterProScan (a sequence analysis application) based on the protein sequence alignment method. InterPro domain database was used. InterPro (http://www.ebiac.uk/interpro/) is a database that integrates diverse information about protein families, domains, and functional sites, and is freely available to public via web-based interfaces and services. Central to the database are diagnostic models, known as signatures, that are used to search protein sequences and investigate their potential functions. InterPro is useful in the large-scale analysis of whole genomes and meta-genomes as well as to characterize individual protein sequences.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation
The KEGG database was used to annotate protein pathways. We used the KEGG online service tool called KEGG Automatic Annotation Server (KAAS) to annotate proteins. The annotation result was mapped on KEGG database online service tool KEGG mapper.
Analysis of subcellular localization
We used WoLF PSORT, a subcellular localization predication software, to predict the subcellular localization of proteins. It is an updated version of PSORT/PSORT II used for the predication of eukaryotic sequences. CELLO, specific for protokaryon species, was used for subcellular localization predication.
GO analysis for functional enrichment
Proteins were classified by GO annotation into 3 categories, namely, biological process, cellular compartment, and molecular function. For each category, a 2-tailed Fisher’s exact test was used to test the enrichment of the differentially expressed proteins and specific proteins against all the identified proteins. GO analysis with a corrected P<0.05 was considered significant.
Pathway enrichment analysis
Encyclopedia of Genes and Genomes database was used with 2-tailed Fisher’s exact test to evaluate the enrichment of differentially expressed proteins and specific proteins against all the identified proteins. The pathway with a corrected P<0.05 was considered significant. These pathways were classified into hierarchical categories according to the KEGG website.
Protein domain enrichment analysis
For each category of proteins, InterPro (a resource that provides functional analysis of protein sequences by classifying them into families and predicting the presence of domains and important sites) database was used and a 2-tailed Fisher’s exact test was employed to investigate the enrichment of differentially expressed proteins and specific proteins against all the identified proteins. Protein domains with a corrected P<0.05 were considered significant.
RESULTS
Characteristics of CE-exo and HD-exo
The results of TEM analysis showed that CE-exo and HD-exo exhibited a round or cup shape in a size range of 50 to 150 nm and were surrounded by a membrane bilayer, consistent with the features of exosomes (Fig. 1A). NTA results revealed a mean diameter of 124.3 nm and a concentration of 9.8E+11 particles/ml for CE-exo, whereas the mean diameter and concentration for HD-exo were 119.0 nm and 1.4E+11 particles/ml, respectively (Fig. 1B, C). Thus, the concentration of CE-exo was higher than that of HD-exo, indicating that patients with CE release more exosomes than healthy individuals.

Characteristics of CE-exo and HD-exo. (A) Transmission electron microscopies of CE-exo and HD-exo. (B) Nanoparticle tracking analysis results showing concentration and diameter of CE-exo and HD-exo. (C) Concentration of CE-exo and HD-exo captured by nanoparticle tracking analysis. Exosomes isolated from the serum of patients infected with cystic echinococcosis. Exosomes isolated from the serum of healthy donors.
Proteomic similarities and differences between CE-exo and HD-exo
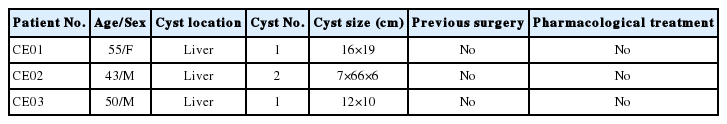
Unstained SDS-PAGE gel showed similar protein content for CE-exo and HD-exo (Fig. 2A). The results of western blot analysis revealed the expression of the typical exosomal markers CD9 and CD63 for both exosome types (Fig. 2B). A total of 373 proteins were identified, and 49 of these were specifically expressed in CE-exo. Among these 49 specific proteins, there were 4 parasitic proteins. Maximum specificity was observed for tubulin alpha-1C chain (XP_024347583.1), a protein present in multiple exosomes [24]. We also found 3 other parasite proteins, including histone H4 (XP_024345389.1) involved in nucleosome formation, Tubulin beta chain (XP_ 024348768.1) involved in the formation of exosomes, and GTP-binding protein SAR1b (XP_024348189.1) involved in the transport of exosomes. There also were Other 45 human-originated proteins exclusively present in CE-exo which may contribute to understanding of the interactions between parasite and host (Supplementary Table S1).
Functional enrichment analysis and subcellular localization of the differentially regulated proteins
The results of proteomic analysis revealed 8 proteins (fold change>1.5) that were differentially regulated in CE-exo. Including 5 proteins that were overexpressed in CE-exo and 3 proteins that were downregulated in CE-exo (Fig. 3A; Table 2). All the 3 downregulated proteins were localized in the extracellular region (Fig. 3B). While, the localization of the 5 upregulated proteins differed (Fig. 3C); for instance, polymeric immunoglobulin receptor (PIGR) was localized in the plasma membrane, while platelet basic protein, immunoglobulin heavy variable 3-30-5, and serum amyloid A-4 protein were present in the extracellular and cytoplasmic regions. GO enrichment analysis of the differentially expressed proteins showed that the top 3 biological processes included “positive regulation of cell division,” “regulation of cell division,” and “cell chemotaxis.” The top 3 cellular components included “whole membrane,” “secretory vesicle,” and “secretory granule” (Fig. 3D). Furthermore, the differentially regulated proteins were enriched in the amyotrophic lateral sclerosis pathway (data not shown).
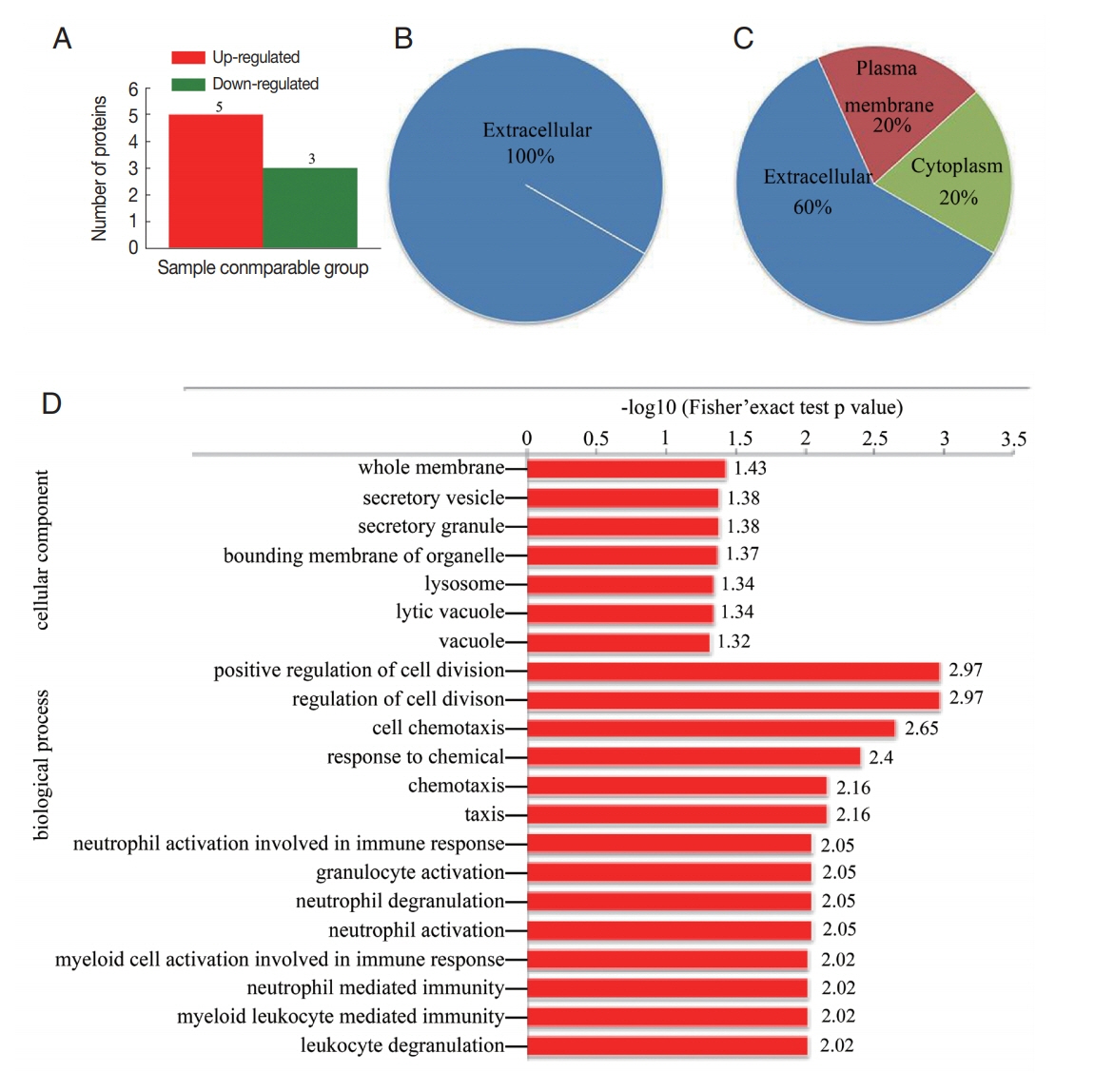
Differentially expressed proteins in CE-exo. (A) Distribution of differentially regulated proteins. (B) Subcellular localization of down-regulated proteins and (C) up-regulated. (D) GO enrichment of differentially regulated proteins.
Enrichment of the proteins exclusively expressed in CE-exo
The enrichment analysis of the proteins exclusively expressed in CE-exo revealed the top 3 biological processes, including “platelet aggregation,” “actin filament-based process,” and “regulated exocytosis”. The top 3 cellular compartments included “actin cytoskeleton,” “extracellular exosome,” and “extracellular vesicle”, while the top 3 molecular functions were “adhesion molecule binding,” “actin binding,” and “cytoskeletal protein binding” (data not shown). Furthermore, KEGG pathway enrichment analysis revealed the proteins that were significantly enriched in 17 pathways (data not shown). The top 5 pathways included “hsa04510 adhesion pathway,” “hsa04810 regulation of actin cytoskeleton pathway,” “hsa04611 platelet activation pathway,” “hsa04530 tight junction pathway,” and “hsa04670 leukocyte transendothelial migration pathway.”
DISCUSSION
CE diagnosis is mainly dependent on microscopical examination and serological tests, which may not detect the disease at early stages. It is noteworthy that the number of confirmed patients with CE is far lower than the number of serologically positive individuals, and the exact reason underlying this observation is unclear. One possibility is that some people may contract E. granulosus once, but the parasites may fail to survival against host immune responses. The related reactions involved in this process are still unknown. Echinococcus is a multicellular parasite, and the interactions between the host and the parasite are complex. The fluid-filled hydatid cyst parasite growing in the host internal organs is surrounded by a 2-layered wall. The inner germinative layer is encompassed by an acellular parasite-derived laminated layer, which exists as a physical barrier. The development of E. granulosus may be accompanied with an interchange of material between the host and the parasite. The release of exosomes by E. granulosus has been recently identified [20], but whether these released exosomes could pass through the thick outer membrane is unknown. In this study, we identified 49 proteins that were exclusively expressed in CE-exo, including 45 huaman proteins and 4 E. granulosus-derived proteins. This observation implies that the exosomes derived from E. granulosus could enter the human circulation and may participate in the communication between E. granulosus and the host. The majority of the 45 human-derived proteins present in CE-exo were associated with cytoskeleton structure, and some of these had anti-microbial activity. These proteins may be expressed in response to stimulation from parasites and may provide a new insight into the interaction between the parasite and the host. The ubiquitous exosomes provided new methods to understand the establishment and development of many diseases and contributed to diagnosis of many diseases [25–27]. In pancreatic cancer, the miRNAs from exosomes serve as powerful tools for early diagnosis, classification, and metastasis monitoring [28]. Exosome-based diagnosis strategies have been developed for lung and ovarian cancers [29]. Nowadays, studies on exosomes isolated from parasites indicated that exosomes not only participated in the interactions between hosts and parasites, but also provided biomarkers for vaccination and diagnosis [30]. Proteomic analysis of exosomes secreted from Schistosome contained protein vaccine candidates [31]. Research on exosomes derived from lymphatic filariasis unearthed biomarkers of stage-specific diagnosis [32]. Study on exosomes in sera from individuals infected with Schistosome provided a new tool for diagnosing schistosomiasis in patients with low parasite burden [33]. Hence, the 4 parasite-derived proteins may contribute as diagnosis candidates and may be useful to improve the accuracy of CE diagnosis.
The concentration of exosomes was shown to increase in response to stress conditions [31]. Patients with diabetes had significantly higher levels of exosomes in their circulation than euglycemic control individuals [34]. The release of exosomes from human umbilical vein endothelial cells significantly increases in response to ischemia [35]. As per our NTA results, the concentration of CE-exo was higher than that of HD-exo in the same amount of (250 μl) serum samples. Both humans and E. granulosus could release exosomes, which may contribute to the excess of exosomes in CE serum samples; however, the validity of this hypothesis and the role played by these exosomes in human body are unclear. More patient samples and further experiments are warranted in future studies.
Aside from these specific proteins, some other proteins were significantly enriched or downregulated in CE-exo. One of the upregulated proteins, PIGR, is a key participant in the formation and secretion of secretory IgA, which is critical for the immune defense against microbial infection and colonization [36]. Studies have demonstrated that the deletion of PIGR results in an increase in the load of Helicobacter pylori [37], Giardia muris [38], and Clostridium [39], and that the sIgA-mediated microbial sensing promoted notovirus and reovirus infection. The role of PIGR in CE is unclear and needs further exploration. The remaining upregulated proteins were involved in protective immune responses such as antigen recognition [40]. Thus, it may be speculated that these upregulated proteins are expressed as a reaction from the host to cystic Echinococcus and may be useful for the treatment and prevention of CE. Histidine-rich-glycoprotein (HRG), one of the downregulated proteins, exerts antibacterial and antifungal activities, and its expression is significantly inhibited in septic mice [41]. Moreover, the supplementary treatment with HRG was shown to improve their survival [42]. Consistent with our results, HRG was significantly downregulated in CE-exo, indicating that HRG may be potentially useful as a candidate molecule for the treatment or vaccination of CE. The other 2 downregulated proteins performed protective functions. Collectin-11 is associated with protection against Schistosomiasis [43], while glutathione peroxidase-3 protects cells and enzymes from oxidative damage [44]. These downregulated proteins may be related with the mechanisms by which the parasite evades the host immune system. These results suggest the new possibilities of treatment and prevention of CE. The exosomes may emerge with dual characters. To further identify the functions of these proteins and exosomes, in vivo and in vitro experiments are deemed necessary.
The 8 differentially regulated proteins were enriched in the amyotrophic lateral sclerosis pathway, but the exact relationship between the pathway and infection is unclear. KEGG pathway enrichment analysis revealed 17 pathways that were related with the proteins exclusively expressed in CE-exo; most of these pathways were relevant to the host response to infections, including cell motility, cell proliferation, cell survival, platelet activation, leukocyte transendothelial migration, and inflammatory suppression, and are related to each other. Thus, the infection with E. granulosus induced changes in our body, and the role of exosomes is important and complex and demands further studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Nature Science Foundation of China (Grant NO. 31560258), Ningxia Natural Science Found Project (NO. 2019AAC03266), and West China First-class Discipline Construction Project in Basic Medicine funded by Ningxia Medical University (NO. 30181602). We would like to thank PTM-Biolabs Co., Ltd. (Hangzhou, China) for the quantitative proteomic analysis assistance.
Notes
CONFLICT OF INTEREST
The authors have declared no conflicts of interest.