Abstract
Cryptosporidium parvum and Giardia duodenalis are the main diarrhea-causing parasitic pathogens; however, their prevalence in Korea is unknown. Here, we conducted a survey to determine the prevalence and genotype distribution of these 2 pathogens causing acute diarrhea in 8,571 patients hospitalized in 17 Regional Institute of Health Environment sites in Korea, during 2013–2016. C. parvum and G. duodenalis were detected and genotyped by nested PCR, and the isolate were molecularly characterized by sequencing the glycoprotein 60 (Gp60) and β-giardin genes, respectively. The overall prevalence of C. parvum and G. duodenalis was 0.37% (n=32) and 0.55% (n=47), respectively, and both pathogens were more prevalent in children under 9 years old. Molecular epidemiological analysis showed that the C. parvum isolates belonged to the IIa family and were subtyped as IIaA13G2R1, IIaA14G2R1, IIaA15G2R1, and IIaA18G3R1. Analysis of the β-giardin gene fragment from G. duodenalis showed that all positive strains belong to assemblage A. This is the first report on the molecular epidemiology and subtyping of C. parvum and G. duodenalis in such a large number of diarrheal patients in Korea. These results highlight the need for continuous monitoring of these zoonotic pathogens and provide a basis for implementing control and prevention strategies. Further, the results might be useful for epidemiological investigation of the source of outbreak.
-
Key words: Cryptosporidium parvum, Giardia duodenalis, glycoprotein 60, β-giardin
The protozoan genera
Cryptosporidium and
Giardia are important causes of diarrhea worldwide [
1,
2]. The main clinical symptoms of infections with
Cryptosporidium spp. include diarrhea, along with vomiting and abdominal cramps, loss of appetite, and low-grade fever. Similarly,
Giardia spp. cause diarrhea, bloating and fatigue, stomach cramps, nausea, and weight loss during chronic infections [
3,
4]. Previous reports have shown that
Giardia duodenalis is the etiological agent in 35.2%, while
Cryptosporidium spp. was the causal agent in 60.3%, of cases among the 199 documented worldwide outbreaks due to waterborne transmission from 2004 to 2010 [
5]. More recently, at least 381 outbreaks of parasitic protozoa were documented globally from January 2011 to December 2016 and the most common etiology was
Cryptosporidium spp., which was responsible for 63% of cases, and
Giardia duodenalis was detected in 37% of cases [
6].
In Korea, a national program to analyze stool samples for the control and prevention of diarrheal diseases revealed a positive rate of
C. parvum and
G. duodenalis of 0.28% and 0.61%, respectively, based on enzyme immunoassays [
7]. In another report,
G. lamblia and
C. parvum were detected by enzyme-linked immunosorbent assay (ELISA) in 2.5% and 0.4% of 6,071 stool samples collected from a general hospital in Gyeonggi province [
8]. Moreover,
Cryptosporidium oocysts and
Giardia cysts have been detected in raw vegetables, environmental soil, and intake water samples [
9,
10]. Two other recent accidental outbreaks of these protozoa in Korea have been reported to date [
11,
12]. The first was a waterborne outbreak in April 2010 including 9 cases of
giardiasis that occurred among individuals drinking valley water stored in a water tank without sterilization as a temporary water supply [
11]. The second was also a waterborne outbreak from May to June 2012 including 124 cases of cryptosporidiosis that originated from an apartment water tank with worn plumbing systems that became contaminated by
Cryptosporidium oocysts [
12]. In Korea, the outbreaks caused by
C. parvum and
G. duodenalis infection are typically accompanied by acute diarrhea; however, surveys of the prevalence of such cases and the causal strains, such as
C. parvum and
G. duodenalis, are rare because the tap water supply system is generally well established throughout the country. Therefore, there has been no molecular epidemiological surveillance to date among the Korean population with respect to acute diarrhea. Accordingly, the aim of the present study was to determine the molecular infection rate and subtypes of
C. parvum and
G. duodenalis responsible for acute diarrhea in Korean patients from 2013 to 2016.
During the 4 years of the study, from January 2013 to December 2016, a total of 8,571 stool samples were collected from acute diarrhea patients in 17 Regional Institute of Health Environment sites located throughout the country. Each stool sample (1 g) was suspended in 5 ml of phosphate-buffered saline (PBS) and filtered through gauze to remove large particles. The filtered liquid was centrifuged at 800 g for 10 min. The supernatant was carefully removed, and the remaining sediment was mixed with 1 ml PBS. The pellet was subjected to 10 boiling (100°C) and freezing (−70°C) cycles to break the hard surface wall of Giardia and Cryptosporidium cysts.
Genomic DNA was extracted from each stool sample using DNAzol (MRC, Cincinnati, Ohio, USA) and stored at −20°C until use. The target DNA for nested PCR was the partial
Cryptosporidium oocyst wall protein (COWP) gene. In the first PCR, a 550-bp fragment was amplified using the Cry-15 (5′-GTAGATAATGGAAGA GATTGT G-3′) and Cry-9 (5′-GGACTGAAATACAGGCATTATCTT G-3′) primers. In the second PCR, a 310-bp fragment was amplified using the cowpnest-F1 (5′-TGTGTTCAATCAGACACAGC-3′) and cowpnest-R2 (5′-TCTGTATATCCTGGTGGGC-3′) primers [
13].
G. duodenalis was detected by nested PCR of the β-giardin gene. The first primer set was G376A forward (5′-CCATCCATAACGACGCCATCGCGGCTCTC-3′) and GGR789-809B reverse (5′-GGC GCT TAG TGC TTT GTG ACC-3′), and the second primer set was G376B (5′-CGA CGC CAT CGC GGC TCT CAG GAA GGA GG-3′) and G759A (5′-CGC CCT GGA TCT TCG AGA CGA CGT CCT-3′), amplifying fragments of 415 bp and 374 bp [
11]. The PCR cycle conditions for the PCR were identical to those used for the primary PCR. PCR products and restriction fragments were subjected to electrophoretic separation on 2% agarose gels, which were stained with Safe-Pinky (GenDEPOT Barker, Texas, USA) and visualized on a gel documentation system (Sangene; G-Box; Cambridge, UK).
The total positive rate of all samples was 0.89%, with a
C. parvum infection rate of 0.37% and a
G. duodenalis infection rate of 0.55%. There were only 2 cases of mixed infection (
Table 1). The positive rates and pattern obtained here using PCR are similar to those reported previously by Cheun et al. [
7] in Korea, using methods other than PCR; however, the rates were lower than those reported in China (1.4%) and in Libya (1%) [
14,
15].
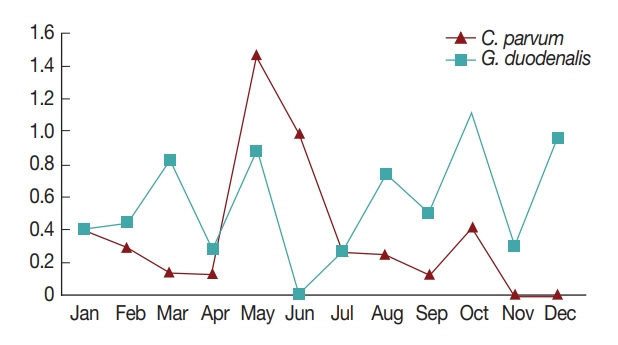
In general, diarrhea caused by infection by waterborne protozoa mainly occurs in the periods between spring and summer, and between summer and fall during the season transition [
7]. Consistently, in the present study, the highest positive rates of the 2 pathogens were observed in May and October (
Fig. 1). This might be due to the change in seasons from spring to summer in May, and from fall to winter in October; however, more research is needed to explain the seasonal nature of protozoan infection [
7,
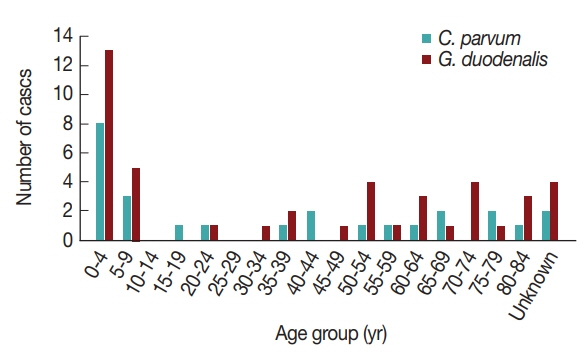
16]. Furthermore, the pathogens were more prevalent in patients under 10 years of age compared to other ages (
Fig. 2). Previously, the infection rate of
G. duodenalis was found to be the highest in the less than 10-year-old group [
17] and to occur most frequently in the summer [
16]. One of the reasons for the high infection rate in children under 10 years of age may be due to their sensitivity to exposure of these pathogens via food and environmental risk factors such as food contamination and accidental ingestion of these organisms [
18]. This general pattern indicates that more attention should be paid to warning young children about the possibility of waterborne pathogen infection.
Interestingly, the positive rates of
C. parvum and
G. duodenalis were very low and did not correlate with increase in age (
Table 1). Although acute diarrhea cases caused by
C. parvum and
G. duodenalis in Korea are rare, these results demonstrate that regular epidemiological surveillance among patients with acute diarrhea could offer guidance for the control of water-borne diseases. Subtyping has been extensively performed in studies of
C. parvum and
G. duodenalis transmission in humans and animals, and nearly 20
C. parvum subtype families and 8
G. duodenalis genetic assemblages have been described at the respective loci [
19,
20]. We conducted phylogenetic analysis of the GP60 gene of
C. parvum using PCR amplification of ~400 base pairs (bp). Nested PCR was performed using the AL3531 forward primer (5′-ATAGTCTCCGCTGTATTC-3′) and AL3533 reverse primer (5′-GAGATATATCTTGGT GCG-3′) and the second set of primers were AL3532 forward primer (5′-TCCGCTGTATTCTCAGCC-3′) and LX0029 reverse primer (5′-CGAACCACATTACAAATGAAGT-3′) [
21]. Nested PCR amplification of β-giardin gene was used to analyze the molecular characteristics of
G. duodenalis. In the primary PCR reaction, a 753 bp fragment was amplified using the G7 forward primer (5′-AAGCCCGACGACCTCACCCGCAGTGC-3′) and G-759 reverse primer (5′-GAGGCCGCCCTGGATCTTCGAGACGAC-3′) and the second set of primers were G511F forward primer (5′-GAACGAACGAGATCGAGGTCC-3′) and G-511R reverse primer (5′-CTCGACGCG CTTCGTGTT-3′) [
22]. All PCR-positive products were purified using a DNA purification kit (Qiagen, Hilden, Germany) after electrophoresis, and then purified again on an agarose gel and sequenced using the ABIPRISM 3730xl Analyzer system (Applied Biosystems, Foster City, California, USA). DNA sequences were compared using the BLAST tool, and phylogenetic trees were constructed using the CLUSTAL W multiple sequence alignment computer program (Histon, Cambridgeshire, UK) and the Molecular Evolutionary Genetics Analysis (MEGA) program with the maximum-likelihood method; the robustness of groupings was assessed using 1,000 bootstrap replicates of the data [
23]. Unfortunately, phylogenetic analysis could only be performed in 47 of the 77 positive samples. A total of 11 samples for
C. parvum and 20 samples for
G. duodenalis were successfully amplified by nested PCR.
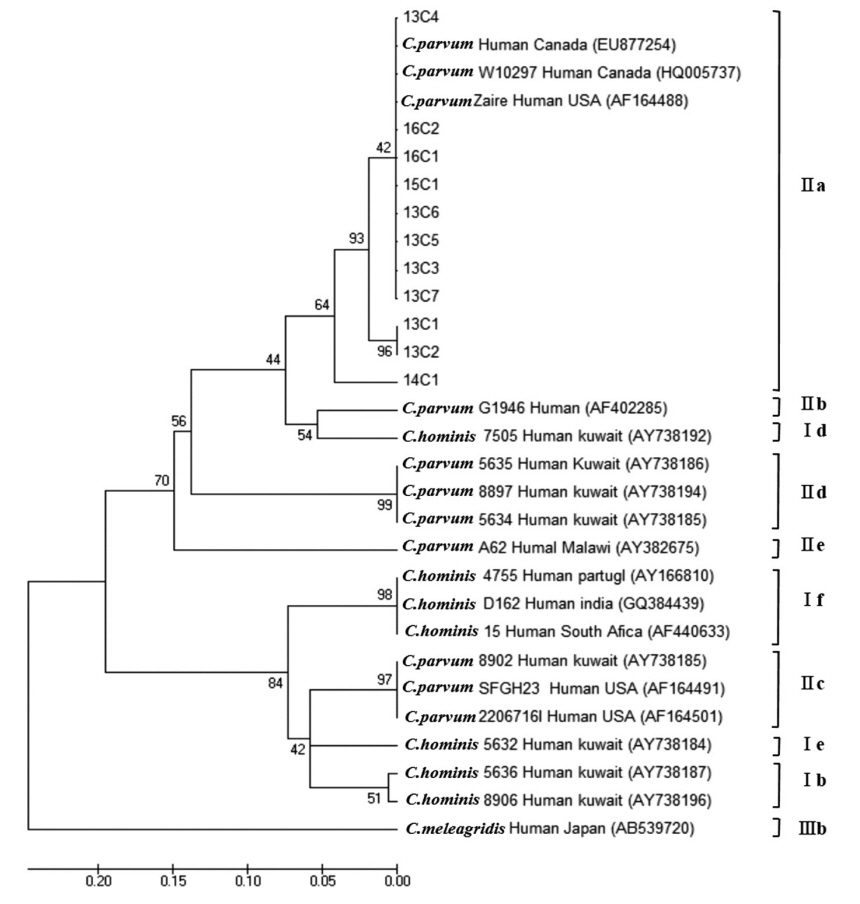
C. parvum was classified into 4 subtype families: 1 in IIaA13G2R1, 7 in IIaA14G2R1, 2 in IIaA15G2R1, and 1 in IIaA18G3R1, representing the most common zoonotic subtype family (
Table 2;
Fig. 3). According to the recent review of genetic diversity in
Cryptosporidium, the IIaA15G2R1 subtype is particularly dominant in calves and lambs as well as in humans; IIaA13G2R1 was reported mostly in calves and AIDS patients; IIaA18G3R1 in cows and in people of Northern Ireland; and IIaA14G2R1 was reported for the first time in cattle from Germany, in AIDS patients from Malaysia and Italy, as well as in fish (Nile tilapia) in Papua New Guinea [
19].
Main source of outbreaks with
Cryptosporidium spp. infection were identified as recreational waters and animal contact, and other sources such as environmental contact, person-to-person spread, food and drinking water supplies comprised a very low proportion [
24]. Especially among the subtypes of
C. parvum, IIaA15G2R1 was the predominant in England and Wales, IIdA24G1 was the most common type in the outbreak in Sweden [
24,
25], and IIaA14G2R1 was identified as dominant in Korea through the present surveillance. With respect to the molecular subtyping analysis, based on these previous reports, we cautiously speculate that the
C. parvum detected from Korean diarrhea stool samples may have originated from contact with animals such as calves, cattle, and lambs raised in Korea. Unfortunately, there was no molecular subtyping conducted in the first outbreak of cryptosporidosis in Korea for source of origin [
12].
Among the 8 genetic assemblages of
Giardia, assemblages A and B are known to be the main causes of human infections [
26]. Although the majority of studies on the inter-species transmission of
G. duodenalis have focused on animal-to-human transmission, there is increasing evidence that
Giardia cysts of human origin can also contaminate the environment and infect wild mammals [
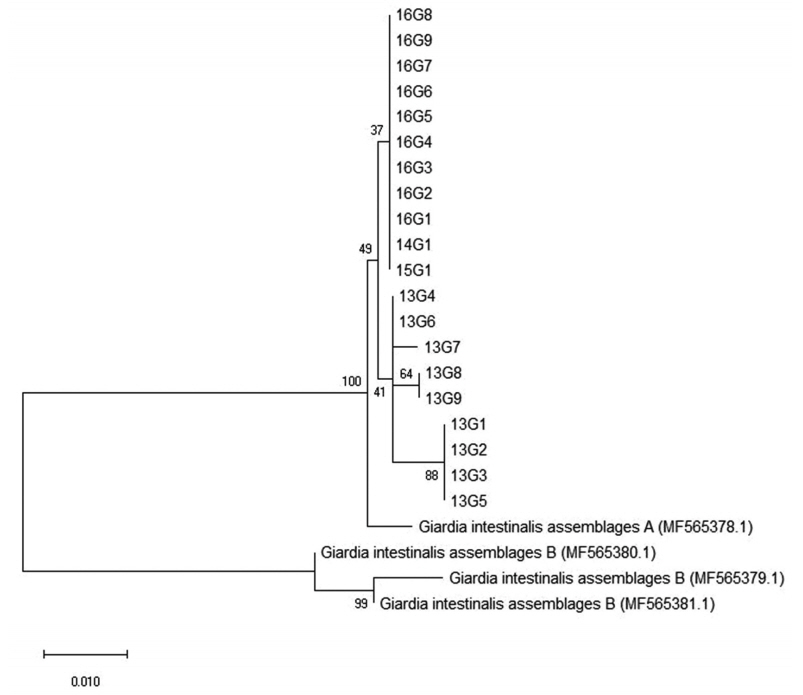
27]. In the present investigation, all positive cases with
G. duodenalis infection were identified to belong to assemblage A (
Table 2;
Fig. 4). This is in line with previous results from the feces DNA of Japanese, Mongolian, and Korean patients [
28,
29]. Because
Giardia cysts have also been detected in natural water resources, it is possible that these patients were naturally infected or exposed to environmental contamination of the pathogen; however, there was no information available on the site of infection or route and timing of exposure. Unfortunately, molecular subtyping was not conducted in the first outbreak of giardiasis in Korea for comparison [
11].
In summary, this is the first molecular epidemiological investigation and subtyping of C. parvum using the Gp60 gene and of G. duodenalis using the β-giardin gene in stool samples from diarrheal patients in Korea. Both pathogens were found to belong to a zoonotic subtype family. These findings emphasize the importance of continuous molecular epidemiological surveillance and subtyping for waterborne parasitic pathogens among acute diarrheal patients to identify the origin of waterborne outbreaks of C. parvum and G. duodenalis infections.
Notes
-
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this study.
ACKNOWLEDGMENT
This work was supported by the Centers for Disease Control and Prevention (4847-311, 2016), Ministry of Health and Welfare, Republic of Korea.
Fig. 1Monthly positive rates of Cryptosporidium parvum and Giardia duodenalis in stool samples of Korean patients with acute diarrhea sampled during 2013–2016.
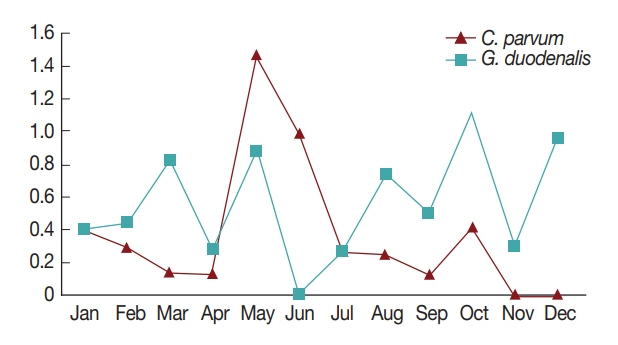
Fig. 2Number of Cryptosporidium parvum and Giardia duodenalis cases by age group in Korea, 2013–2016.
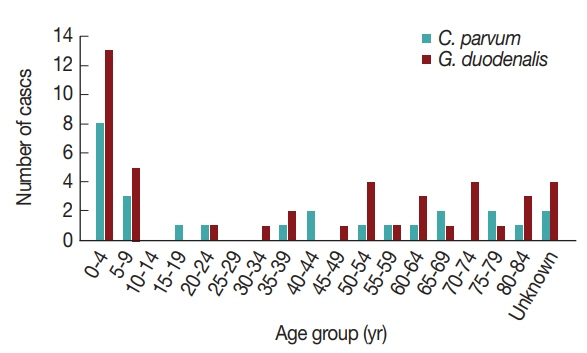
Fig. 3Phylogenetic relationships of the GP60 locus of Cryptosporidium species strains isolated from Korean patients with diarrhea. The phylogenetic tree was constructed using the maximum-likelihood method with 1,000 replicates (implemented using Molecular Evolutionary Genetics Analysis [MEGA]). Other Cryptosporidium species sequences were obtained from GenBank.
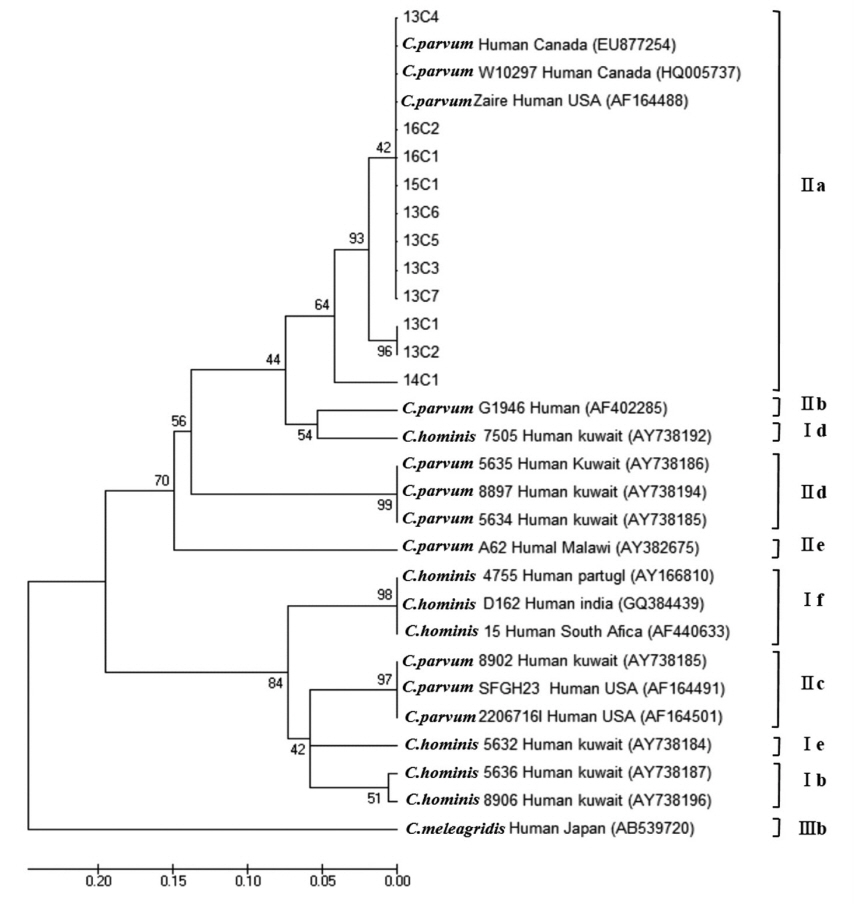
Fig. 4Phylogenetic relationships of the β-giardin locus of Giardia species strains isolated from Korean patients with diarrhea. The phylogenetic tree was constructed using the maximum-likelihood method with 1,000 replicates (implemented using Molecular Evolutionary Genetics Analysis [MEGA]). Other Giardia species sequences were obtained from GenBank.
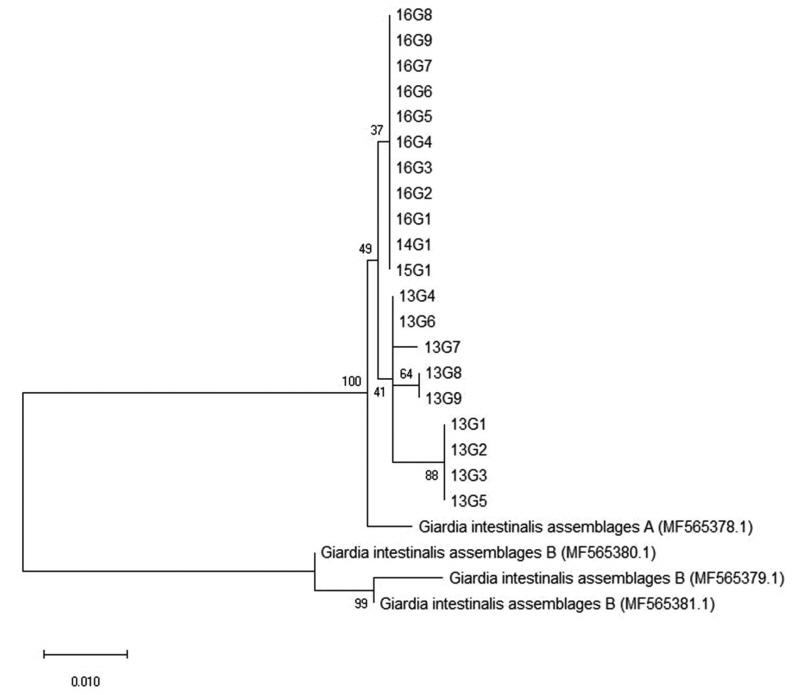
Table 1Detection of Cryptosporidium parvum and Giardia duodenalis infections using PCR in human fecal samples in 2013–2016
Table 1
|
Year |
No. of sample |
Positive no. of C. parvum (%) |
Positive no. of G. duodenalis (%) |
|
2013 |
3,093 |
23 (0.74) |
27 (0.87) |
|
2014 |
2,046 |
2 (0.1) |
9 (0.44) |
|
2015 |
1,722 |
2 (0.12) |
1 (0.06) |
|
2016 |
1,710 |
5 (0.29) |
10 (0.68) |
|
Total |
8,571 |
32 (0.37) |
47 (0.55) |
Table 2Subtyping of the GP60 genes and genotyping of the β-giardin for human stool sample of diarrheal patients from Korea testing positive for Cryptosporidium parvum and Giardia duodenalis using nested PCR
Table 2
|
Specimen ID |
Subtypes (GP60) |
|
Specimen ID |
Genotypes (β-giardin) |
|
C. parvum
|
13C1 |
IIaA14G2R1 |
G. duodenalis
|
13G1 |
Assemblage A |
|
13C2 |
IIaA14G2R1 |
|
13G2 |
Assemblage A |
|
13C3 |
IIaA14G2R1 |
|
13G3 |
Assemblage A |
|
13C4 |
IIaA14G2R1 |
|
13G4 |
Assemblage A |
|
13C5 |
IIaA14G2R1 |
|
13G5 |
Assemblage A |
|
13C6 |
IIaA14G2R1 |
|
13G6 |
Assemblage A |
|
13C7 |
IIaA14G2R1 |
|
13G7 |
Assemblage A |
|
14C1 |
IIaA13G2R1 |
|
13G8 |
Assemblage A |
|
15C1 |
IIaA18G3R1 |
|
13G9 |
Assemblage A |
|
16C1 |
IIaA15G2R1 |
|
14G1 |
Assemblage A |
|
16C2 |
IIaA15G2R1 |
|
15G1 |
Assemblage A |
|
|
|
|
16G1 |
Assemblage A |
|
|
|
|
16G2 |
Assemblage A |
|
|
|
|
16G3 |
Assemblage A |
|
|
|
|
16G4 |
Assemblage A |
|
|
|
|
16G5 |
Assemblage A |
|
|
|
|
16G6 |
Assemblage A |
|
|
|
|
16G7 |
Assemblage A |
|
|
|
|
16G8 |
Assemblage A |
|
|
|
|
16G9 |
Assemblage A |
References
- 1. Cacciò SM, Thompson RC, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol 2005;21:430-437.
- 2. Sánchez A, Munoz M, Gómez N, Tabares J, Segura L, Salazar Á, Restrepo C, Ruíz M, Reyes P, Qian Y, Xiao L, López MC, Ramírez JD. Molecular epidemiology of Giardia, blastocystis, and Cryptosporidium among indigenous children from the Colombian Amazon basin. Front Microbiol 2017;8:248.
- 3. Moore CE, Elwin K, Phot N, Seng C, Mao S, Suy K, Kumar V, Nader J, Bousfield R, Perera S, Bailey JW, Beeching NJ, Day NP, Parry CM, Chalmers RM. Molecular characterization of Cryptosporidium species and Giardia duodenalis from symptomatic Cambodian children. PLoS Negl Trop Dis 2016;10:e0004822.
- 4. Homan WL, Mank TG. Human giardiasis: genotype linked differences in clinical symptomatology. Int J Parasitol 2001;31:822-826.
- 5. Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2004–2010. Water Res 2011;45:6603-6614.
- 6. Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks-An update 2011–2016. Water Res 2017;114:14-22.
- 7. Cheun HI, Cho SH, Lee JH, Lim YY, Jeon JH, Yu JR, Kim TS, Lee WJ, Cho SH, Lee DY, Park MS, Jeong HS, Chen DS, Ji YM, Kwon MH. Infection status of hospitalized diarrheal patients with gastrointestinal protozoa, bacteria, and viruses in the Republic of Korea. Korean J Parasitol 2010;48:113-120.
- 8. Huh S, Moon SG, Lim YH. A survey of intestinal protozoan infections among gastroenteritis patients during a 3-year period (2004–2006) in Gyeonggi-do (province), South Korea. Korean J Parasitol 2009;47:303-305.
- 9. Sim S, Won J, Kim JW, Kim K, Park WY, Yu JR. Simultaneous molecular detection of Cryptosporidium and Cyclospora from raw vegetables in Korea. Korean J Parasitol 2017;55:137-142.
- 10. Lee MY, Cho EJ, Lee JH, Han SH, Park YS. A survey of Cryptosporidium oocysts in water supplies during a 10-year period (2000–2009) in Seoul. Korean J Parasitol 2010;48:219-224.
- 11. Cheun HI, Kim CH, Cho SH, Ma DW, Goo BL, Na MS, Youn SK, Lee WJ. The first outbreak of giardiasis with drinking water in Korea. Osong Public Health Res Perspect 2013;4:89-92.
- 12. Cho EJ, Yang JY, Lee ES, Kim SC, Cha SY, Kim ST, Lee MH, Han SH, Park YS. A waterborne outbreak and detection of Cryptosporidium oocysts in drinking water of an older high-rise apartment complex in Seoul. Korean J Parasitol 2013;51:461-466.
- 13. Yu JR, Lee SU, Park WY. Comparative sensitivity of PCR primer sets for detection of Cryptosporidium parvum
. Korean J Parasitol 2009;47:293-297.
- 14. Wang T, Fan Y, Koehler AV, Ma G, Li T, Hu M, Gasser RB. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol 2017;51:127-131.
- 15. Gashout A, Taweni F, Elmabrouk H. Pattern of intestinal parasites among hospital patients at Tripoli Central Hospital, Libya. Libyan J Med Sci 2017;1:13-15.
- 16. Yoder JS, Harral C, Beach MJ. Giardiasis surveillance-United States, 2006–2008. MMWR Surveill Summ 2010;59:15-25.
- 17. Seguí R, Muñoz-Antoli C, Klisiowicz DR, Oishi CY, Köster PC, de Lucio A, Hernández-de-Mingo M, Puente P, Toledo R, Esteban JG, Carmena D. Prevalence of intestinal parasites, with emphasis on the molecular epidemiology of Giardia duodenalis and Blsstocystis sp., in the Paranaguá Bay, Brazil; a community survey. Parasit Vectors 2018;11:490.
- 18. Sockett PN, Rodgers FG. Enteric and foodborne disease in children: A review of the influence of food- and environment-related risk factors. Paediatr Child Health 2001;6:203-209.
- 19. Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium
. Trends Parasitol 2018;34:997-1011.
- 20. Heyworth MF.
Giardia duodenalis genetic assemblages and hosts. Parasite 2016;23:13.
- 21. Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 2005;43:2805-2809.
- 22. Huey CS, Mahdy MA, Al-Mekhlafi HM, Nasr NA, Lim YA, Mahmud R, Surin J. Multilocus genotyping of Giardia duodenalis in Malaysia. Infect Genet Evol 2013;17:269-276.
- 23. Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 2008;9:299-306.
- 24. Chalmers RM, Robinson G, Elwin K, Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors 2019;12:95.
- 25. Gherasim A, Lebbad M, Insulander M, Decraene V, Kling A, Hjertqvist M, Wallensten A. Two geographically separated food-borne outbreaks in Sweden linked by an unusual Cryptosporidium parvum subtype, October 2010. Euro Surveill 2012;17:20318.
- 26. Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 2011;24:110-140.
- 27. Hogan JN, Miller WA, Cranfield MR, Ramer J, Hassell J, Noheri JB, Conrad PA, Gilardi KV.
Giardia in mountain gorillas (Gorilla beringei beringei), forest buffalo (Syncerus caffer), and domestic cattle in Volcanoes National Park, Rwanda. J Wild Dis 2014;50:21-30.
- 28. Matsubayashi M, Kimata I, Abe N. Identification of genotypes of Giardia intestinalis isolates from a human and calf in Japan. J Vet Med Sci 2005;67:337-340.
- 29. Hong SH, Anu D, Jeong YI, Abmed D, Cho SH, Lee WJ, Lee SE. Molecular characterization of Giardia duodenalis and Cryptosporidium parvum in fecal samples of individuals in Mongolia. Am J Trop Med Hygiene 2014;90:4-47.