Partial Characterization of Two Cathepsin D Family Aspartic Peptidases of Clonorchis sinensis
Article information
Abstract
Cathepsin D (CatD, EC 3.4.23.5) is a member belonging to the subfamily of aspartic endopeptidases, which are classified into the MEROPS clan AA, family A1. Helminth parasites express a large set of different peptidases that play pivotal roles in parasite biology and pathophysiology. However, CatD is less well known than the other classes of peptidases in terms of biochemical properties and biological functions. In this study, we identified 2 novel CatDs (CsCatD1 and CsCatD2) of Clonorchis sinensis and partially characterized their properties. Both CsCatDs represent typical enzymes sharing amino acid residues and motifs that are tightly conserved in the CatD superfamily of proteins. Both CsCatDs showed similar patterns of expression in different developmental stages of C. sinensis, but CsCatD2 was also expressed in metacercariae. CsCatD2 was mainly expressed in the intestines and eggs of C. sinensis. Sera obtained from rats experimentally infected with C. sinensis reacted with recombinant CsCatD2 beginning 2 weeks after infection and the antibody titers were gradually increased by maturation of the parasite. Structural analysis of CsCatD2 revealed a bilobed enzyme structure consisting of 2 antiparallel β-sheet domains packed against each other forming a homodimeric structure. These results suggested a plausible biological role of CsCatD2 in the nutrition and reproduction of parasite and its potential utility as a serodiagnostic antigen in clonorchiasis.
INTRODUCTION
Clonorchis sinensis is a fish-borne trematode parasite, which causes clonorchiasis in humans. The parasite is prevalent in Asian countries including China, Korea and northern Vietnam, with an estimated 35 million people infected worldwide [1]. Chronic infection with the parasite induces periductal inflammation, fibrosis, cholangitis, cholelithiasis, and cholangiectasis [1–3]. Strong epidemiological correlations between clonorchiasis and the incidence of cholangiocarcinoma suggest that C. sinensis is a group I biological carcinogen that can induces or facilitates cholangiocarcinoma in humans [4].
Cathepsin D (CatD; also known as aspartic peptidase), carrying 2 catalytic aspartate residues in the active site, belongs to the peptidase family A1 of the MEROPS clan AA [5]. This clan includes several subfamily enzymes such as CatD (EC 3.4.23.5), pepsin (EC 3.4.23.1), chymosin (EC 3.4.23.4), and renin (EC 3.4.23.15). CatD is less well known than other types of peptidases in terms of biological function and abundance in parasitic helminths [6,7]. The enzymes have been reported to initialize the degradation of host hemoglobin and trigger molecular pathogenesis in blood-feeding helminths, and therefore, the CatDs of helminth parasites are of great interest as targets for potential vaccine or therapeutic drugs [8–12]. To our knowledge, however, there are no studies investigating CatD or its homologs in C. sinensis.
In this study, we report the identification and partial characterization of 2 novel CatDs of C. sinensis (CsCatDs). The 2 CsCatDs were expressed at various developmental stages of C. sinensis. CsCatD2 was specifically synthesized in the parasite’s eggs and intestine. The antigenic property of CsCatD2 also suggested its diagnostic value as a serodiagnostic antigen in clonorchiasis.
MATERIALS AND METHODS
Parasite
C. sinensis metacercariae were collected from naturally infected intermediate host, Pseudorasbora parva, in Korea. Sprague-Dawley rats were infected by oral feeding with 100 metacercariae. The 2-week-old juvenile worms and 4-, 6-, and 9-week-old adult worms were harvested from the livers of the rats after infection as described previously [13,14]. The worms were washed 5 times with cold physiological saline to remove any contamination from the hosts and then were used immediately for RNA preparation for cDNA synthesis or stored at −70°C until use. The cDNA was prepared from C. sinensis worms according to the same method described previously [13,14].
Cloning of genes encoding 2 CsCatDs
The nucleotide sequences of 2 CsCatDs, named CsCatD1 and CsCatD2, were identified during expressed sequence tags (EST) analysis of the cDNA library of C. sinensis adult worms [15]. The homology patterns of the ESTs were analyzed against the non-redundant database by using the BLASTX program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The full-length genes for 2 CsCatDs were amplified from C. sinensis cDNA by polymerase chain reaction (PCR) using the primers flanking the open reading frame (ORF) of each gene. The forward and reverse primers for CsCatD1 were 5′-ATGATTCATCTGGGCTTGTTGTTTTGG-3′ and 5′-TCACCATCCGAATCCGAACAATCTGGA-3′. For CsCatD2, 5′-ATGCGATTTTACGCCATCTTGCTGCTT-3′ and 5′-CTAAGTGGACCTTGCAAAGCCAACACG-3′ were used. The PCR product was analyzed on 1.2% agarose gel, gel-purified and ligated into the T&A cloning vector (Real Biotech Corporation, Banqiao City, Taiwan). The ligated plasmid DNA was transformed into Escherichia coli DH5α competent cells (Real Biotech Corporation) and positive clones were selected by colony PCR. The nucleotide sequence of each cloned gene was analyzed by automated DNA sequencing. Nucleotide sequences of CsCatD1 and CsCatD2 were deposited to GenBank database under accession numbers of GU433604 and GU433605, respectively.
Analysis of sequence features of CsCatDs
Primary amino acid sequences of CsCatDs were deduced from the nucleotide sequences using LASERGENE software package (DNASTAR, Madison, Wisconsin, USA). Physico-chemical properties and molecular weight were analyzed using ProtScale (http://www.expasy.org/tools/protscale.html) and the ExPASy ProtParam Tool (http://web.expasy.org/protparam/), respectively. N-terminal signal peptide, N-glycosylation site were predicted using SignalP v4.1 [16] and NetNGlyc v1 (http://www.cbs.dtu.dk/services/NetNGlyc/), respectively.
Phylogenetic tree construction
The phylogenetic tree was constructed using the neighbor-joining method with MEGA4 (http://www.megasoftware.net). Bootstrap proportions were used to assess the robustness of the tree with 1,000 bootstrap replications.
Transcriptional profile of 2 CsCatDs across developmental stages of C. sinensis
Expressions of 2 CsCatDs in different developmental stages of C. sinensis were analyzed by semi-quantitative reverse transcription PCR (RT-PCR) with 5 μg of each total cDNA, which were prepared from each developmental stage, including metacercariae, 2-week-old juveniles, and 4-, 6-, and 9-week-old adults, according to the previous same method [13,14]. The specific primers used for RT-PCR were the same primers described above. The C. sinensis β-actin gene (GenBank ID: EU109284.1) was also amplified as an internal control. The amplicons were analyzed on 1.2% agarose gel and observed under ultraviolet (UV).
Expression and purification of recombinant CsCatDs (rCsCatDs)
To produce rCsCatDs, fragment deleting the signal peptide region was amplified from each gene by PCR. For CsCatD1, 5′-GAGCTCGTTATTCGGATTCCTCTAATCGGA-3′ and 5′-GTCGACTCACCATCCGAATCCGAACAATCT-3′, which contained a 5′ Sac I site and 5′ Sal I site, were used. Two primers, 5′-GGATCCAAAGTTTTGAGAGTTCCGCTCAAA-3′ and 5′-GTCGACCTAAGTGGACCTTGCAAAGCCAAC-3′, which harbored a 5′ BamH I site and 5′ Sal I site, were used for CsCatD2. Each amplified PCR product was subcloned into the T&A cloning vector (Real Biotech Corporation) and was transformed into E. coli DH5α. The resulting plasmid DNA was digested, ligated into the pQE-30 expression vector (Qiagen, Hilden, Germany), and then transformed into E. coil M15 [pREP4] cells (Qiagen). Expression of recombinant protein was induced by addition of isopropyl-1-thio-β-galactopyranoside (IPTG) to 1 mM final concentration for 3 hr at 37°C with shaking at 200 rpm for aeration. The cells were harvested by centrifugation at 12,000 rpm for 15 min and suspended in 8 M urea lysis buffer. The recombinant protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) chromatography (Qiagen) followed by the protocols provided by the manufacturer. The purification and purity of the recombinant protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Production of polyclonal antibody for rCsCatD2 (anti-CsCatD2)
Polyclonal antibody against rCsCatD2 (anti-CsCatD2) was produced by immunizing mice with the purified rCsCatD2. The protein (100 μg) was mixed with Freund’s adjuvant (Sigma, St. Louis, MO, USA) and intraperitoneally injected into mice 3 times at 2-week intervals. Two weeks after the final booster, the mice were sacrificed and the sera were collected. The immunoglobulin G (IgG) fraction was further isolated from the sera with a Protein G-Sepharose column (Amersham Biosciences, Piscataway, New Jersey, USA) followed by manufacturer’s protocols. The specificity of the anti-CsCatD2 was confirmed by immunoblot. In brief, the purified rCsCatD2 was separated by 12% SDS-PAGE and transferred onto nitrocellulose membrane (Millipore, Billerica, Massachusetts, USA). The membrane was blocked with 3% skim milk in phosphate buffered saline (PBS) for 2 hr at room temperature, washed with PBS containing 0.05% Tween 20 (PBST) 3 times with 10 min intervals. The membrane was then incubated with anti-CsCatD2 (1:1,000 dilutions in PBST) for 2 hr at room temperature. After several washes with PBST, the membrane was incubated with 1:1,000 diluted horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Sigma) for 2 hr at room temperature. The immuno-reactive bands were visualized with 4-chloro-1-naphthol (4CN, Sigma), and the reaction was stopped by washing the membrane with distilled water.
Immunofluorescence assay (IFA)
To investigate the localization of CsCatD2 in C. sinensis adult worm, IFA was performed. The freshly prepared adult worms (6-week old) were washed with PBS (20 mM, pH 7.4) and fixed in 4% paraformaldehyde. The worms were dehydrated with a graded ethanol series, embedded in paraffin blocks and stored in a desiccator until use. The 4-μm-thickness sections were mounted on slide glasses, deparaffinized, rehydrated and rinsed with PBS. The slides were incubated with 1:200 diluted anti-CsCatD2 at room temperature for 2 hr and washed with PBS several times. Normal mouse serum was applied as a negative control [13,17]. After incubation with tetramethylrhodamine isothiocyanate (TRITC)-conjugated anti-mouse IgG (Sigma) diluted 1:200 for 2 hr, the slides were washed with PBS several times and observed using a confocal laser scanning microscope FV-1000 (Olympus, Tokyo, Japan).
Antigenic property analysis of CsCatD2
Time-course antibody production specific for CsCatD2 in rats experimentally infected with C. sinensis was analyzed. Four rats were infected with 100 C. sinensis metacercariae and the sera from each rat were collected at 0, 2, 4, 6, 9, and 12 weeks post-infection. The purified rCsCatD2 was separated on 12% SDS-PAGE, transferred onto nitrocellulose membrane. The membrane cut into strips and blocked with PBST supplemented with 3% skim milk for 1 hr. The strips were then incubated with a 1: 200 dilution of sera from each rat collected at the different time points, respectively, at room temperature for 3 hr. After several washes with PBST, the strips were incubated with 1:1,000 diluted HRP-conjugated anti-rat IgG (Sigma). The immuno-reactive bands were visualized with 4CN (Sigma), and the reaction was stopped by washing the strips with distilled water.
Homology modeling for structure analysis of CsCatD2
The default protocol of YASARA Structure v18.4.24 [18] was used to construct the 3-dimensional (3D) homology models of CsCatD2 using the ‘hm_build.mcr’ macro file (http://www.yasara.org/hm_build.mcr). To construct the homology model, the PSI-BLAST [19] was conducted against PDB entries (updated November, 2018) [20]. Two experimentally resolved structures of CatDs derived from the tick Ixodes ricinus (PDB ID: 5N7N, [21]) and humans (PDB ID:4OD9, [22]) were used as templates. The homology model was built on each template and refined using high-resolution energy minimization of YASARA force field [23]. A hybrid homology model was selected by combining the best scoring components of the reliable models for zymogen and mature form. Dimerization of the structure was predicted using GalaxyGemini [24] and the potential binding energies of the homodimers were calculated using the PISA program [25]. A positive free energy of dissociation ΔGdiss indicates chemically stable associations. All graphic structures were prepared using the open source code of PyMOL (The PyMOL Molecular Graphics System, Version 1.7.2.1, Schrödinger, LLC) under Linux and CHIMERA v8.6.1 [26]. Interfacial residues between the 2 chains were analyzed using ‘InterfaceResidues’ python script (https://pymolwiki.org/index.php/InterfaceResidues) in PyMOL v1.7.2.1. Overall structural variations were measured and visualized using ENDscript v2 [27] with the PDBAA95 database, E-value of 1e-12 and contact range of 2.7 Å.
RESULTS
Sequence and phylogenetic analyses of CsCatDs
The full-length genes encoding CsCatD1 and CsCatD2 were identified. The gene encoding CsCatD1 spanned 1,278 bp in length encoding a protein of 424 amino acids, with a predicted size of 46.6 kDa. The ORF of CsCatD2 comprises 1,158 bp, which encodes an approximately 42.5 kDa protein containing 385 amino acids. Multiple sequence analysis of the 2 CsCatDs suggested that the proteins shared similar primary structures with previously identified CatDs (Fig. 1A). All sequences aligned revealed high positional conservation in the functional residues and elements of CatD family enzymes; however, the N-terminal pro-domain region and C-terminal region differed from each other. The 2 aspartic acid residues, which act as catalytic residues in that active site of CatD family enzymes, were well conserved in CsCatD1 (Asp87 and Asp273) and CaCatD2 (Asp86 and Asp272). Both CsCatDs contained the predicted signal peptide sequences in their N-termini. The Y flap, one of the essential elements underlying the activity of CatD family enzymes, and key residues associated with the formation of binding pocket in the active site were well conserved in both CsCatDs. Four predicted N-glycosylation sites (Asn45, Asn54, Asn124, and Asn215) were identified in CsCatD1, but CsCatD2 carried only a single potential N-glycosylation site at Asn323. The CsCatD1 revealed a high level of amino acid sequence identity (more than 50%) with all the sequences analyzed, especially 89.9% with CatD of Opisthorchis viverrini (OvCatD). Meanwhile, the CsCatD2 showed lower levels of sequence identity with other CatDs including OvCatD (52.0%), Fasciola hepatica (FhCatD; 49.1%), Schistosoma mansoni (SmCatD; 51.0%) and S. japonicum (SjCatD; 51.7%). Two CsCatDs showed 53.3% identity with each other. The phylogenetic analysis of 2 CsCatDs showing identity with CatD family of enzymes from other organisms revealed that CsCatDs were clustered into a clade with CatDs of trematode parasites (Fig. 1B).
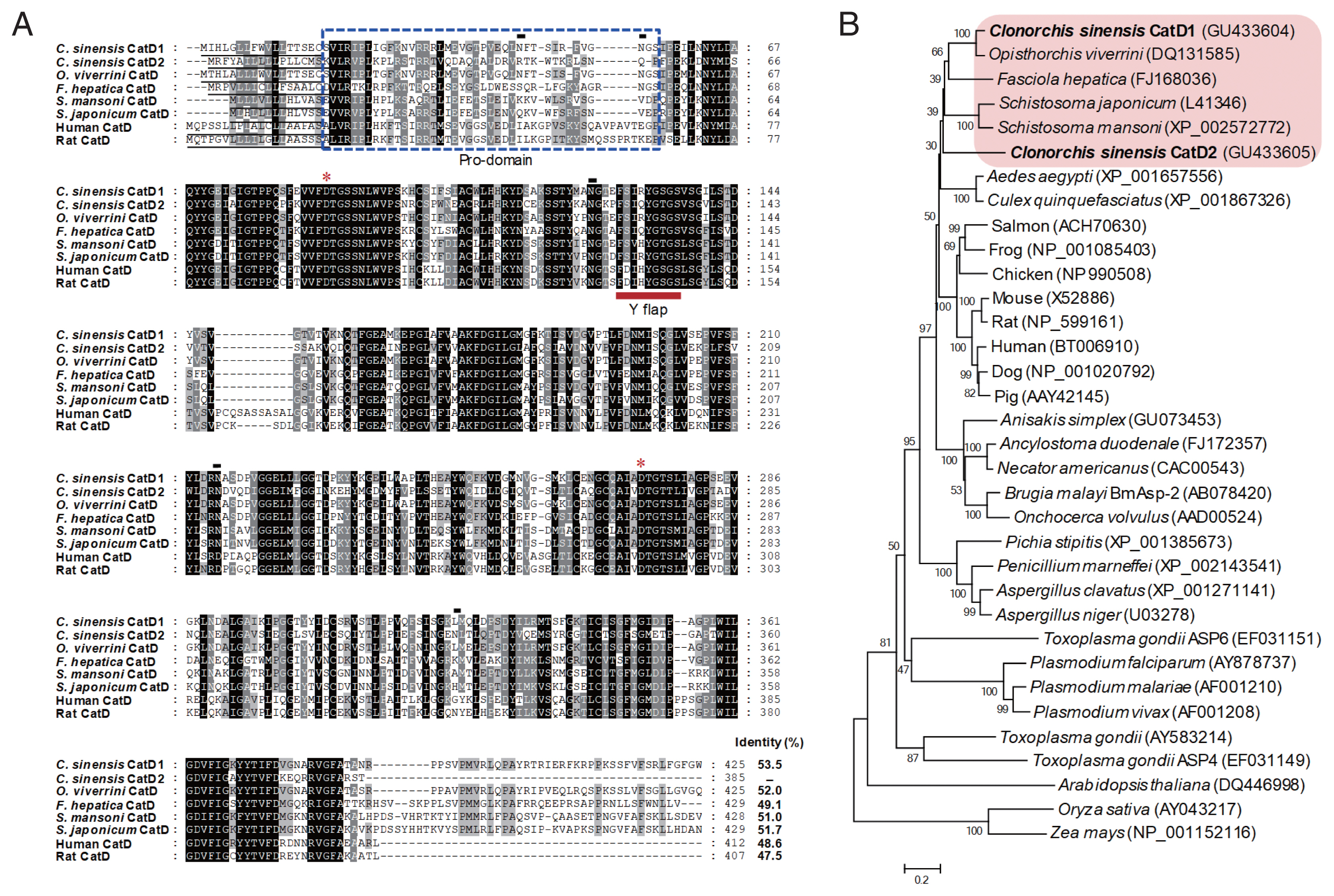
Multiple sequence alignment and phylogenetic analyses. (A) Multiple sequence analysis. The deduced amino acid sequences of 2 CsCatDs were aligned with those of Opisthorchis viverrini (AAZ39883), Fasciola hepatica (ABJ97284), Schistosoma mansoni (AAB63442), Schistosoma japonicum (AAC37302), Human (NP_001900), and Rat (NP_599161). Gaps are introduced into the sequences to maximize alignment. The asterisks indicate the catalytic aspartic acid residues. Black lines under the sequences are predicted signal peptide sequences. Potential pro-peptide regions are marked with a blue dotted box. The Y flap element is indicated by a bold red line. Predicted N-glycosylation sites in CsCatDs are represented by black lines. The shading represents the degree of identity among the sequences: black (100%), dark grey (50–92%), and light grey (15–49%). (B) Phylogenetic analysis. The phylogenetic tree was constructed with the neighbor-joining method using the MEGA4 program. Numbers on the branches indicate bootstrap proportions (1,000 replicates).
Expression profiles of CsCatDs in the developmental stages of C. sinensis
The expression profiles of 2 CsCatDs were analyzed via semi-quantitative RT-PCR in different developmental stages of C. sinensis. CsCatD1 was expressed at different developmental stages, juveniles (2 weeks) and adult worms (4, 6, and 9 weeks), and the expression was gradually increased by maturation of the parasite (Fig. 2). CsCatD2 also showed similar patterns of expression; however, it was expressed at all developmental stages of the parasites analyzed, from metacercariae to adults (Fig. 2).
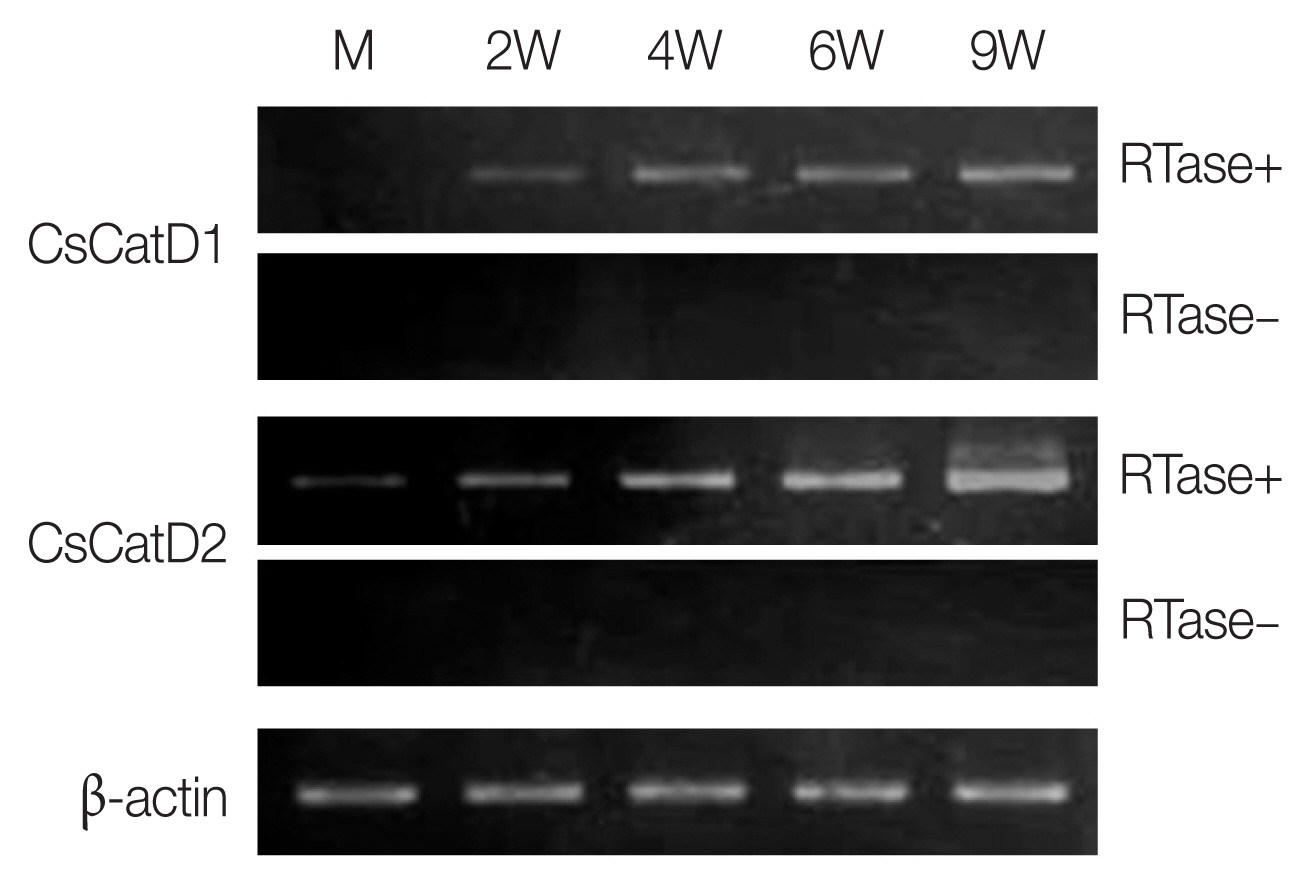
Expression profiles of CsCatDs in different developmental stages of C. sinensis. Transcriptional profiles of CsCatDs at various developmental stages of C. sinensis were analyzed via semi-quantitative RT-PCR. Semi-quantitative RT-PCR was performed as described in Materials and Methods. The reaction was conducted in the presence (+) or absence (−) of reverse transcriptase (RTase) to verify DNA contamination. C. sinensis actin gene was amplified as an internal control. M, metacercariae; 2W, 2-week-old juvenile worms; 4W, 4-week-old adult worms; 6W, 6-week-old adult worms; 9W, 9-week-old adult worms.
Localization and antigenic properties of CsCatD2
The rCsCatD2 was expressed in E. coli as an insoluble form with an estimated size consistent with the predicted molecular size of about 42 kDa (Fig. 3A). However, rCsCatD1 was not successfully expressed in E. coli. Therefore, only CsCatD2 was analyzed further in this study. A polyclonal antibody against rCsCatD2 (anti-CsCatD2) was generated in mice and its specificity was confirmed by immunoblot (Fig. 3A). IFA was performed to determine the localization of CsCatD2 in C. sinensis adult worms. Positive fluorescence signals were strongly detected in the epithelial cells lining the intestine and in the eggs within the uterus of the parasite (Fig. 3B). The antigenic property of CsCatD2 was also analyzed by immunoblot. CsCatD2 reacted with the sera from rats infected with C. sinensis. Specific antibodies of CsCatD2 were detected in the sera from all 4 rats and the antibody titers were gradually increased by time-dependent infection (Fig. 3C). In the early stage of infection (2 weeks), antibodies against CsCatD2 were detected only in 1 rat, but the antibodies were produced in all 4 rats at 6 weeks post-infection.
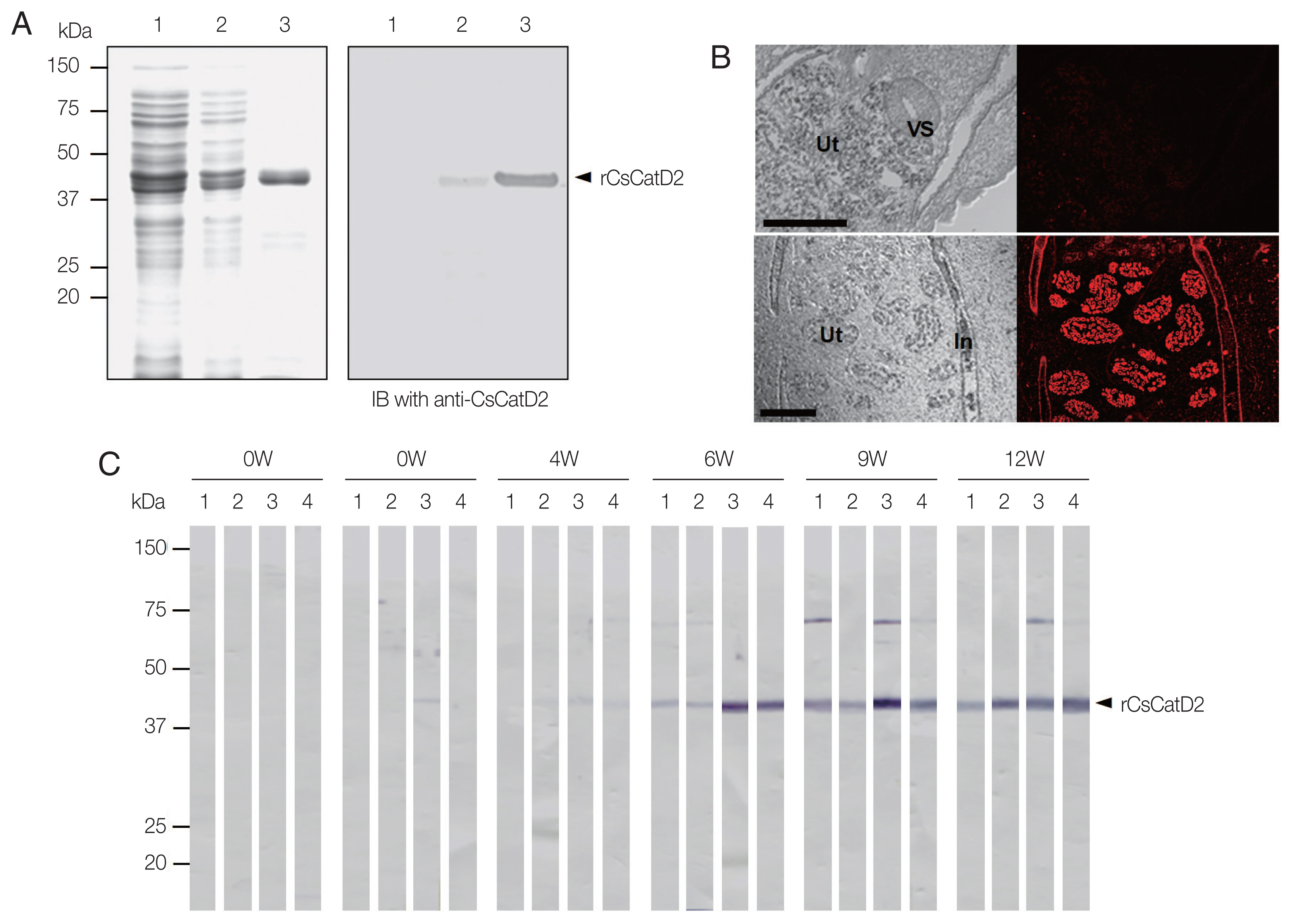
Expression and characterization of CsCatD2. (A) Productions of rCsCatD2 and anti-CsCatD2. The rCsCatD2 was produced as an insoluble protein in E. coli. Proteins were analyzed by SDS-PAGE and stained with Coomassie blue (left). Anti-CsCatD2 was produced by immunizing mice with purified rCsCatD2. The specificity of anti-CsCatD2 was confirmed by immunoblot (right). Lane 1, Escherichia coli lysate control; lane 2, IPTG-induced E. coli lysate; lane 3, affinity-purified rCsCatD2. (B) Localization of CsCatD2. IFA was performed with anti-CsCatD2 against sections of C. sinensis adult worms. The slide was observed with a confocal laser scanning microscope (×10 magnification). Scale bar indicated 100 mm. (C) Antigenic property of CsCatD2. The time course of antibodies targeting CsCatD2 in rats experimentally infected with C. sinensis was analyzed. Four rats were infected with 100 C. sinensis metacercariae and the sera from each rat were collected at 0 (0W), 2 (2W), 4 (4W), 6 (6W), 9 (9W), and 12 (12W) weeks post-infection. The purified rCsCatD2 was separated on 12% SDS–PAGE, transferred onto nitrocellulose membrane, cut into strips, and then probed with sera from four rats.
Structural analysis of CsCatD2 by homology modeling
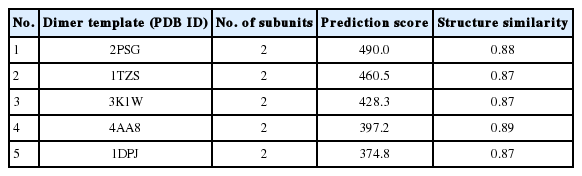
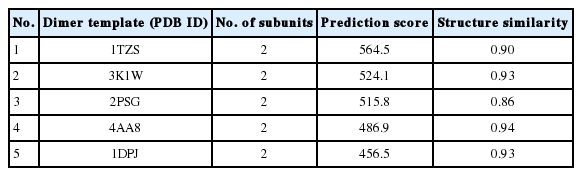
The zymogen and mature CsCatD2 models were designed to delineate the structural features of CsCatD2. Zymogen and mature forms of CsCatD2 structure were predicted using experimentally-determined structures of CatDs derived from I. ricinus (PDB ID: 5N7N) and humans (PDB ID: 4OD9), respectively. The 2 structures were further validated as suitable models. The zymogen and mature forms of CsCatD2 were predicted to form homodimers (Fig. 4), as supported by the predicted structural similarity for potential homodimerization of each monomer using GalaxyGemini (Supplementary Tables S1, S2). The interface areas of the homodimeric assembly for zymogen and mature forms were 3,035 Å2 and 4,294 Å2, respectively, suggesting that approximately 20% to 30% of the overall accessible surface area of the 2 interacting monomers was involved in the formation of homodimeric assembly. Free energy of dimer dissociation yielded ΔGdiss values of −27.6 kcal/mol and −24.1 kcal/mol for zymogen and mature forms, respectively. Both zymogen and mature CsCatD2 models revealed well-conserved bilobed structures, comprising 2 antiparallel β-sheet domains packed against each other (Fig. 5A). The interdomain links between the N- and C-terminals contribute to close localization of 2 catalytic aspartic acid residues, Asp86 and Asp272, to constitute the active site pocket. The CsCatD2 zymogen model contained a pro-peptide structure consisting of a conserved part A (light navy) and a variable part B (green). When the pro-peptide region was eliminated from the zymogen, the N-terminal region of the mature enzyme was predicted to turn toward β-sheet (pink) from the unstructured loop in zymogen (Fig. 5A). The Y flap region (cyan) and the polyproline loop (purple) appear to cover the pocket of the substrate-binding site. Gly132 was located as a flap tip residue and Met350 was situated as a hinge residue on the opposite side. Analysis of the superposed structure of a total of 120 CatD homologs suggested that the proximity of the active site pocket was highly conserved, but both pro-peptides of zymogen and N-terminal regions of mature enzyme varied relatively between CatD family enzymes (Fig. 5B). Three disulfide bridges, which may stabilize CsCatD2 structure, were predicted.
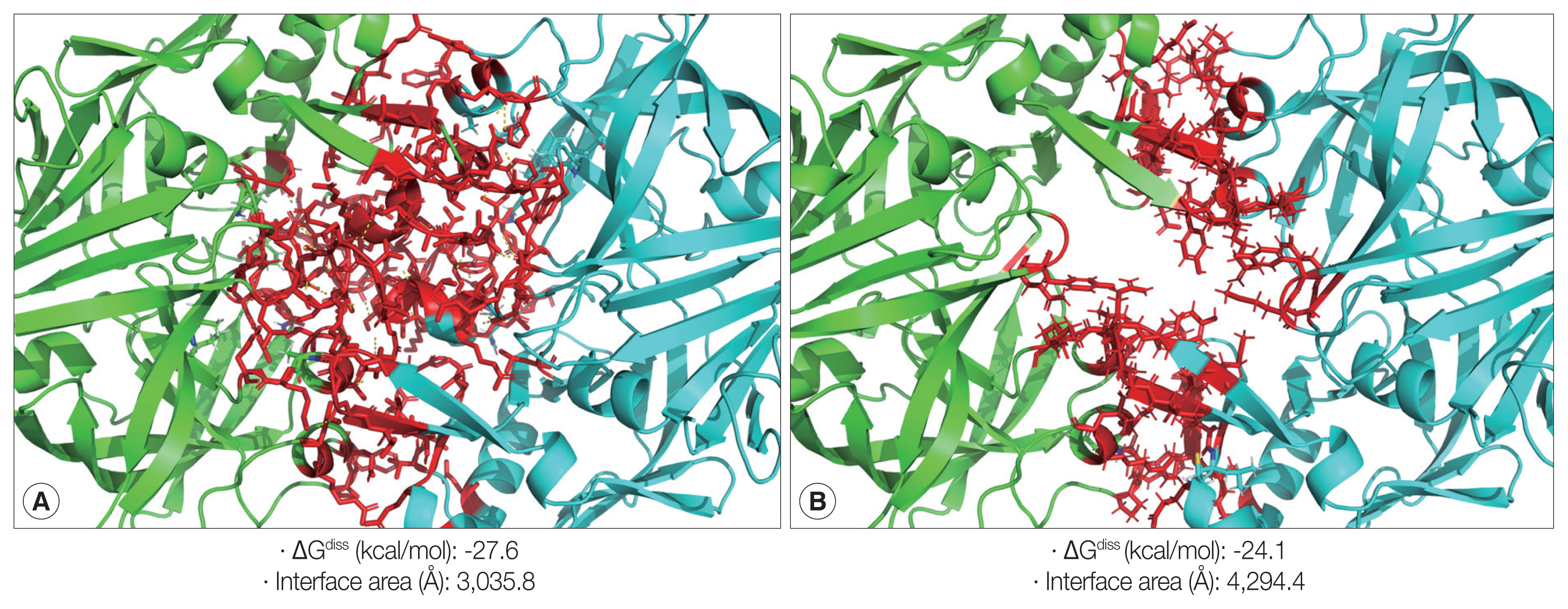
Homodimeric structures of zymogen and mature CsCatD2. The 3D homology modeling analysis of zymogen (A) and mature form (B) of CsCatD2 predicted homodimeric structures of the proteins. The 3D structures of zymogen and mature form of CsCatD2 were predicted based on experimentally resolved structures of CatDs derived from tick Ixodes ricinus (PDB ID: 5N7N) and humans (PDB ID: 4OD9), respectively. The free energies of dissociation (ΔGdiss; kcal/mol), and interface area (Å2) were calculated using the PISA program [25]. The interfacial residues are shown in red sticks, computed using the ‘InterfaceResidues’ script in PyMOL v1.7.2.1 while the remaining structures of chain A (green) and chain B (cyan) are represented as a cartoon.
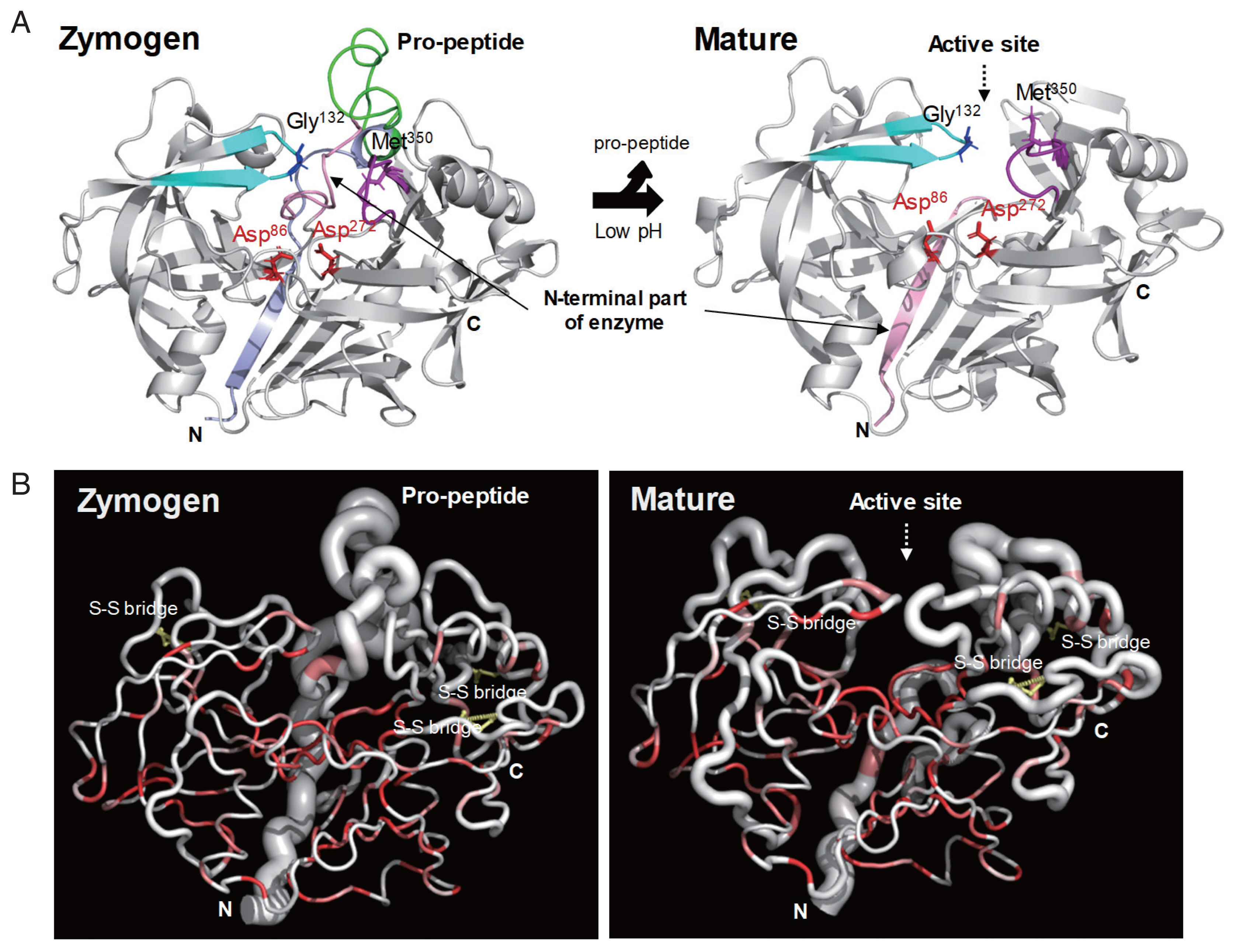
Overall structural features of CsCatD2. (A) Overall 3D structure and secondary structural features of zymogen (A) and mature (B) forms of CsCatD2. The pro-peptide region in zymogen represented as conserved part A (light navy ribbon) and variable part B (green). The Y flap region (cyan) and the polyproline loop (purple) along with the 2 catalytic aspartic residues (red stick) are also marked. The flap tip residue and hinge residue of the polyproline loop are indicated by black and purple sticks, respectively. (B) Sausage representation of the CsCatD2 with sequence and structural conservation rendering. To visualize sequence conservation within the CaCatD2 and CatD family proteins (PDB entries 5UX4, 5N7N, 5N7Q, 5N70, 1TZS, 5MLG, 5MKT, 5NFG, 3D91, 2I4Q, 5T4S, 3PSG, 2PSG, 5PEP, 1PSA, 1F34, 4AA9, 3PEP, 4PEP, 1FLH, 1G0V, 1DP5, 2JXR, 1B5F, 3OAD, 3F9Q, 1M43, 5YIA, 5YID, 1SME, 1LF3, 5BWY, 1PFZ, 3QRV, 2BJU, 1QS8, 2ANL, 1LS5, 1MIQ, 1UH7, 3LIZ, 3FNS, 1YG9, 4RLD, 3QVC, 1LYW, 4OBZ, 3ZLQ, 2EWY, 3ZKM, 6FGY, 2QZL, 1TQF, 3EXO, 3QI1, 2Q11, 3TPJ, 1WKR, 1FKN, 2QK5, 3KMX, 3DM6, 2HM1, 2ZJN, 3IXK, 4DPF, 3L58, 2ZHR, 2FDP, 4TRZ, 4B1D, 1SGZ, 3UDH, 3U6A, 5HTZ, 2ZJK, 2OF0, 3VV6, 3CIB, 5MBW, 1YM2, 1W50, 4L7G, 2QU2, 3CKP, 3HVG, 5MXD, 5CLM, 1YM4, 2Q15, 2ZJI, 5EZX, 2VIE, 3LPI, 4YBI, 2WJO, 3TPR, 5V0N, 2HIZ, 6EJ2, 6EJ3, 2ZJJ, 2VA5, 4B78, 3BRA, 4B70, 4EWO, 2ZJH, 1MPP, 2QZX, 4YBF, 2RMP, 3R1G, 1HTR, 4OBZ, 1LYA, 3OAD, 3APR, 1PSO, and 1CZI), the sausage was colored from white (similarity score below 0.7) to red (strict identity). The degree of structural conservation is proportional to the thickness of the sausage, which corresponds to the mean root-mean-square deviation (RMSD) per residue between Cα pairs based on structural alignments. Putative disulfide bridges are indicated by a yellow line. All the graphics were prepared using ENDscript v2 [27] and PyMOL v1.7.2.1.
DISCUSSION
Helminth parasites express a large set of different classes of peptidases. These enzymes play important roles in the parasite physiology and pathobiology, and therefore represent attractive targets for the development of vaccines or therapeutic agents [17,28–31]. Similar to other helminth parasites, C. sinensis also secretes a large number of different peptidases [32–34]. The multigene family of C. sinensis cathepsin Fs (CsCFs) is the most extensively studied family of peptidases in C. sinensis playing a critical role in parasite biology. CsCFs are expressed in the intestinal epithelium of C. sinensis, actively secreted into the intestinal lumen of the parasite, and are participated in host protein hydrolysis [13,14,35]. Two leucine aminopeptidases of C. sinensis (CsLAP1 and CsLAP2) are also synthesized in the intestine of the parasite and may mediate the final step in the catabolism of host proteins absorbed from the intestine [36]. Asparaginyl endopeptidase of C. sinensis (CsAEP) is another peptidase that is specifically synthesized in the parasite’s intestine [37]. All of these enzymes are expressed in diverse developmental stages of C. sinensis and show maximum levels of expression in the adult stage of the parasite. These findings collectively suggest that these enzymes may be involved in the cooperative digestion of host proteins absorbed by the parasite [14,36,37].
In this study, we identified 2 novel CsCatDs and partially characterized their properties. Both CsCatDs are typical CatD family enzymes that share similar structural features with previously known CatD superfamily proteins. We have expressed the 2 CsCatDs in E. coli, but only CsCatD2 was successfully expressed in E. coli. Although rCsCatD2 was expressed in E. coli, the recombinant protein was produced as an insoluble protein. We have tried refolding of the purified rCsCatD2 under diverse refolding conditions to transform the protein into soluble and functionally active enzyme but, unfortunately, we failed to obtain an enzymatically active protein. Two CsCatDs were expressed in diverse developmental stages of C. sinensis. Although the expression patterns were not completely matched, the levels of both CsCatDs were gradually increased by maturation of the parasite. CsCatD2 was expressed in the intestine of the parasite, suggesting a possible role in the intestinal hydrolysis of host proteins. However, unlike other gut-associated peptidases of C. sinensis including CsCFs, CsLAPs, and CsAEP, which are specifically localized in the intestine of the parasite, CsCatD2 was also highly expressed in eggs, indicating a plausible role in egg production or maturation in the parasite. Our finding that CsCatD2 was also expressed in the intestine of C. sinensis suggests host protein digestion via functional networking of diverse classes of peptidases including CsCFs, CsLAPs, CsAEP, and CsCatD2 in the intestine of the parasite. However, further studies are warranted to elucidate the biological roles of CsCatD2. The time course production of specific antibodies against CsCatD2 in rats experimentally infected with C. sinensis suggests that the enzyme may induce immune response. Antibodies produced in the early stages of infection are assumed to have been produced by intestinal CsCatD2, secreted by the juvenile worms. The increasing pattern of antibodies specific to CsCatD2 observed as the parasite mature suggests that the antibodies may also have been produced against CsCatD2 in eggs produced by the adult worms. These also suggest a potential role of CsCatD2 as a serodiagnostic antigen in clonorchiasis, but further studies are necessary to validate antigenicity of CsCatD2 in human cases and cross-reactivity with other helminth infections.
Structural analysis of CsCatD2 via homology 3D modeling elucidated the structural features of the enzyme. As expected, CsCatD2 showed a high degree of similarity in 3D structure with CatD superfamily of enzymes. Similar to canonical CatD superfamily [38], zymogen and mature CsCatD2 were bilobed with a large substrate-binding crevice between 2 lobes. The conserved signature DTG motif occurring as a single copy in each lobe includes the catalytic aspartic acid residues at positions 86 and 272, which were identified in CsCatD2. Some peptidases in clan AA form homodimers or oligomers that contribute to the overall protein stability [39]. The negative values of free energy dissociation (ΔGdiss) in both zymogen and mature CsCatD2 models suggested that the enzyme may form a homodimeric structure. The active site pocket of CsCatD2 is covered by an antiparallel β-sheet (commonly known as Y flap region), which plays an important role in substrate binding. The flap is a functionally crucial fragment that facilitates substrate binding in the active site pocket and interaction with a ligand or inhibitor.
In conclusion, the 2 CsCatDs are typical enzymes belong to the CatD superfamily. The biological functions of the CsCatDs are not clear but, considering their expression profiles and localization in C. sinensis, they may play a key role in parasite’s nutrition and reproduction. Further studies are necessary to elucidate their biological role.
Supplementary Data
ACKNOWLEDGMENT
This work was supported by a grant of the National Research Foundation (NRF) funded by the Korean government (NRF 2016R1C1B1009348).
Notes
CONFLICT OF INTEREST
The authors declare that they have no competing interests.