Abstract
The third stage larvae (L3) of Anisakis typica were detected in 2 species of threadfin bream, Nemipterus hexodon and N. japonicus, from the Gulf of Thailand, and were morphologically and molecularly characterized. Total 100 threadfin breams, 50 Nemipterus hexodon and 50 N. japonicus, were examined with naked eyes after the opening of abdominal cavity with scissors. Almost all infected larvae remained alive and active even the fish were transported for 1–2 days. Anisakid larvae were exclusively distributed in the body cavity and rarely in the liver. The prevalence of A. typica L3 were 68.0% and 60.0% in N. hexodon and N. japonicus and their infection intensities were 3.5 and 4.2 per fish infected each. Morphological and morphometric analysis were performed by viewing specimens under both a light microscope and a scanning electron microscope. Interestingly, the protruded mucron of Anisakis typica under SEM showed a distinct cylindrical shape that differed from the cone shape of A. simplex. The protruded mucron could be used to identify A. typica L3 larvae in the future. A comparison of the ITS1-5.8S-ITS2 rDNA nucleotide sequences of these species revealed high blast scores with A. typica. Conclusively, it was confirmed that A. typica L3 are prevalent in threadfin breams from the Gulf of Thailand, and their morphological and molecular characters are something different from those of other anisakid larvae, including A. simplex and A. pegreffii.
-
Key words: Nemipterus hexodon, Nemipterus japonicus, Anisakis typica, Ornate threadfin bream, Japanese threadfin bream
INTRODUCTION
In Thailand, various marine species are important natural sources of nutrition, and imperative to industries surrounding the harvest of marine species for maintaining food and economic stability. Previous reports have stated that the demersal resources have declined around 8% between the years 1961 and 1995. Four major species groups in decline include
Nemipterus spp.,
Priacanthus spp.,
Saurida spp. and squids [
1]. Threadfin bream (
Nemipterus spp.), in the order Perciformes, is one of the types of fish in decline. One possible cause may be infection with an endoparasite that causes abnormal fish growth.
Anisakidae is a family in the phylum Nematoda, which is probably the most prevalent family of parasites found in marine mammals. The family has the potential to affect the human health. The intermediate host of anisakid nematodes are marine vertebrates and invertebrates including fish, squid, crustaceans and molluscs. Accidental ingestion of anisakid L3 larvae (e.g.
Anisakis simplex and
Anisakis pegreffii) by humans can cause anisakiasis and more than 20,000 cases have been reported worldwide, over 90% from Japan, and the rest from Asia, Europe, America and New Zealand. Anisakid larvae burrow into the stomach wall and elicit an inflammatory response which causes acute ulceration, nausea, vomiting and epigastric pain [
2–
5]. The histological studies of rat stomach tissue infected by
Anisakis typica found epithelial damage, bleeding within the submucosal layer, inflammation of the stomach and decreased numbers of mucous cap cells, which comprise the gland-containing, mucus-membrane layer covering the surface of the stomach [
6]. However, there are no records of human anisakiasis caused by
A. typica, possibly because there is little information about the infection available.
There have been many reports of cases of anisakiasis in Asia and Europe, especially in Japan, which has popularised cuisine made with raw or undercooked seafood such as sushi and sashimi [
7–
10]. In Thailand, which also has a high level of consumption of raw seafood, eating raw fish or squid with dipping sauce or cooked into spicy seafood salad is common. Today, seafoods transportation from the Gulf of Thailand to the northern areas can be done within a short time and still fresh. So, the chances of becoming infected with the anisakid larvae are increasing as well.
Identification of L3 anisakid larvae morphologically using a light microscope is rather difficult, because of the low level of development of organs at this stage and the lack of remarkable characteristics of the larvae. Consequently, morphological study using a scanning electron microscope (SEM) is required to provide adequate detail, especially with regard to the skin and miniscule structures which are not observable using a light microscope. Moreover, molecular methods, such as DNA sequencing are the most reliable method and are often used to identify anisakid larvae. There have been many reports of anisakid nematodes using internal transcribed spacer (ITS) region of rDNA analysis, such as
Anisakis typica [
11–
17],
Anisakis simplex [
11,
14,
16],
Anisakis pegreffii [
14,
16,
17],
Anisakis brevispiculata, Anisakis paggiae and
Anisakis physeteris [
17].
The purpose of this study was to investigate the epidemiology of anisakid larvae present within marine fish, N. hexodon and N. japonicus, sent from the Gulf of Thailand to Chiang Mai province, northern Thailand, by using both morphological and molecular studies to increase the reliability of the species identification.
MATERIALS AND METHODS
Parasite collection
Total 100 threadfin breams, 50 ornate threadfin bream (
Nemipterus hexodon) and 50 Japanese threadfin bream (
Nemipterus japonicus) (
Fig. 1) purchased from a department store in Chiang Mai province of northern Thailand between December 2018 and June 2019. The fish nomenclature adopted has been outlined in a journal [
18]. Purchased fish were shipped from the Gulf of Thailand, Chachoengsao province (
Fig. 2) under refrigerated conditions (−4°C) for 1–2 days. Almost all of the anisakid larvae were active while carefully removed from the body cavity and liver. Total 243 anisakid larvae, 118 from
N. hexodon and 125 from
N. japonicus were washed using saline (0.85% NaCl) for further study.
Permanent slide preparation and light microscopy
The 30 larvae (15 samples from each fish species) were fixed with 4% formalin for 24 hr, washed with distilled water and stained with hematoxylin for 2 weeks. After staining, samples were dehydrated using a graded alcohol series, were cleared with xylene and mounted in permount. The permanent slides were illustrated using a light microscope with a drawing tube. Measurements were performed using an ocular micrometer and all measurements have been given in micrometers (μm) unless otherwise indicated. The species were identified based on morphology according information previously provided in journals and textbooks [
19–
23].
Scanning electron microscopy
The 18 larvae (9 samples from each fish species) were deep cleaned with normal saline (0.85% NaCl), washed with phosphate buffer and pre-fixed in 2.5% glutaraldehyde. After 24 hr, samples were post-fixed in 1% osmium tetroxide (OsO4) in phosphate buffer for 24 hr, dehydrated using an ethanol series and acetone, were dried using the critical point method with CO2 and mounted with carbon tape on aluminum stubs. The specimens were sputter-coated with gold and examined using a Jeol JSM-5910 LV (Japan) at an accelerating voltage of 15 kV. All measurements have been given in micrometers unless otherwise indicated.
Molecular analyses
Isolation of genomic DNA
The 6 larvae (3 samples from each fish species) were used for extraction of the genomic DNA with 150 μl of 5% Chelex (Fluka) solution containing 3 μl of proteinase K (Sigma). Samples were then heated at 55°C for 1 hr, followed by gentle vortexing and heating at 95°C for 30 min and gently vortexed. The mixture was centrifuged at 13,000 g for 10 sec. The supernatant was removed and stored at −20°C for further use.
DNA amplification and sequencing
PCR amplification of the ribosomal DNA region comprising of the ITS1-5.8s-ITS2 plus 70 nucleotides of the 28S gene was performed using primers NC5 (forward; 5′-GTAGGTGAACCTGCGGAAGGATCATT-3′) and NC2 (reverse; 5′ TTAGTTTC-TTTTCCTCCGCT-3′) [
11]. The PCR amplifications were carried out in a final volume of 20 μl, which included 50 ng of DNA template, 50 pM of each primer (NC5 and NC2), 1.5 mM MgCl
2, 200 μM of each dNTP, and 0.5 unit of
Taq DNA polymerase. The amplification procedure involved an initial denaturation step at 94°C for 5 min, then 30 cycles that included denaturation at 94°C for 30 sec, primer annealing at 60°C for 30 sec, extension at 72°C for 30 sec, and a final extension at 72°C for 5 min. PCR products were analysed after a 100 V, 45 min electrophoretic separation through a 2.0% agarose gel stained with RedSafe nucleic acid staining solution in TBE buffer. Gels were visualized using a UV-transilluminator. PCR products were purified and sequenced by Ward Medic Ltd., Part. (Bangkok, Thailand).
Phylogenetic analysis
The sequences of ITS1-5.8S-ITS2 of rDNA was verified by forward and reverse comparisons which were assembled and edited using the BioEdit Sequence Alignment Editor version 7.0.5.3. They were compared manually with original chromatograms when necessary. The obtained sequences were aligned with previously characterized sequences of Anisakis spp. registered in GenBank using ClustalW in Mega version 6.06. Sequences identified included Anisakis simplex, Anisakis typica, Anisakis typica var. indonesiensis, Anisakis brevisculata, Anisakis nascettii, Anisakis paggiae, Anisakis pegreffii, Anisakis physeteris and Anisakis ziphidarum. Hysterothylacium aduncum was used as an outgroup. Phylogenetic analysis was carried out by maximum likelihood (ML) and neighbour-joining (NJ) methods using bootstrapping of 1,000 replicates in Mega version 6.06.
RESULTS
Prevalence, intensity and abundance of the L3 anisakid larvae
The average length and weight of sampled fish have been shown in
Table 1.
N. hexodon was significantly longer and heavier than
N. japonicus (
P=0.05). Anisakid larvae in
N. hexodon were all in the body cavity, while the distribution in
N. japonicus showed that larvae were mostly in the body cavity and some were in the liver. Morphological characteristics of all anisakid nematodes from 2 species of the fish revealed the same species when investigated under a light microscope. Prevalence, intensity and abundance of larvae in
N. hexodon were 68%, 3.47 and 2.36, and 60%, 4.17 and 2.50, respectively, for
N. japonicus.
The body of L3 anisakid larvae under a light microscope was cylindrical and attenuated at both ends. Measured larvae ranged from 12.31–18.00 (15.78±2.82) mm in length and 0.34–0.45 (0.39±0.04) mm in width. The anterior extremity of each larval specimen examined contained a projecting boring tooth and inconspicuous lips. Esophagus was characterized by an anterior part (proventriculus) with cylindrical striated muscle that was 1.36–1.88 (1.58±0.19) mm in length and 0.08–0.12 (0.10±0.02) mm in width, and a glandular ventriculus that was 0.55–0.94 (0.67±0.13) mm long and 0.10–0.20 (0.15±0.04) mm wide with an oblique esophago-intestinal junction. The rectum of an anisakid larva is surrounded by rectal glands and opens through an anal opening (
Table 2;
Fig. 3). The scanning electron micrograph showed that the characteristic triradiate mouth opening was surrounded by 4, small, cephalic papillae (2 were dorsolateral and 2 were ventrolateral). Each larval specimen had an excretory pore that opened at the base of the ventrolateral lips below its boring tooth. The larvae were covered with a rigid cuticle that was approximately 5 μm thick, with an annular transverse striation and longitudinal lateral grooves (
Fig. 4). Their tails are short and rounded with terminal mucrons. Protrude mucrons were cylindrical-shaped measuring 12.9–18.9 (15.7±3.0) μm long and 6.4–8.8 (7.6± 1.7) μm wide (
Figs. 4,
5). The comparison of the mucron from SEM micrograph of
A. simplex was cone-shape measuring 14.7 μm long and 9.6 μm wide [
24].
A total of 910 bp of ITS1-5.8S-ITS2 of rDNA gene sequences of the present anisakid larvae from
N. hexodon and
N. japonicus were deposited in the GenBank under the accession numbers MN420660 and MN420659, respectively. A comparison of the nucleotide sequences of the rDNA of these species revealed high blast scores with
A. typica accession numbers KC928262, HF911524, JX523715, and AB432909 in the GenBank (percent identity=100). The results of the genetic distance study (
Table 3) showed that the distances between
A. typica from the present study with other
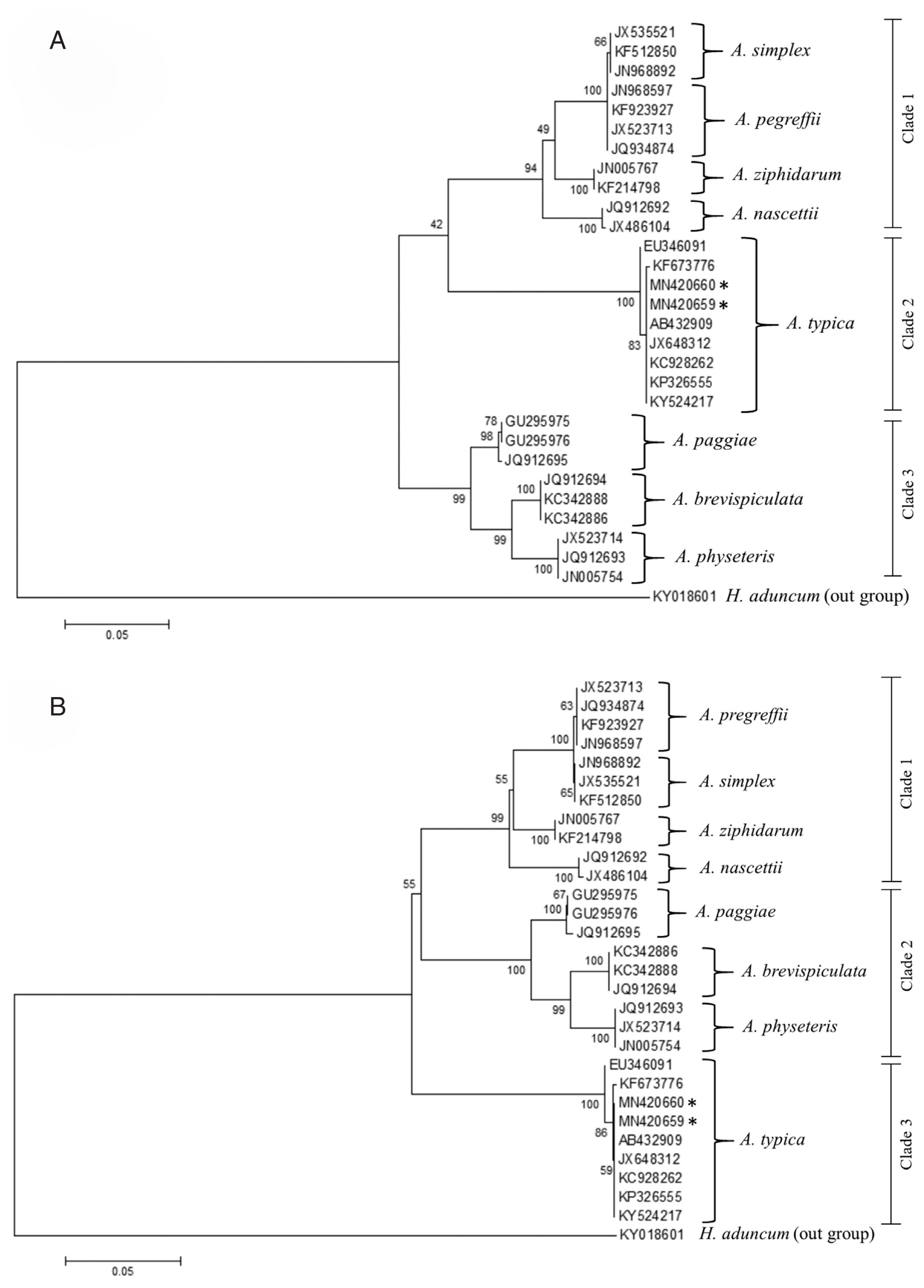
A. typica in the GenBank were 0.000 or 0%. Moreover, the phylogenetic tree of 8
Anisakis species constructed with ML and NJ methods (
Fig. 6) could be divided into 3 major clades and anisakid larvae in the present study were grouped together with other
A. typica with 100% bootstrap value. The phylogenetic tree constructed using ML and NJ methods showed that the group of
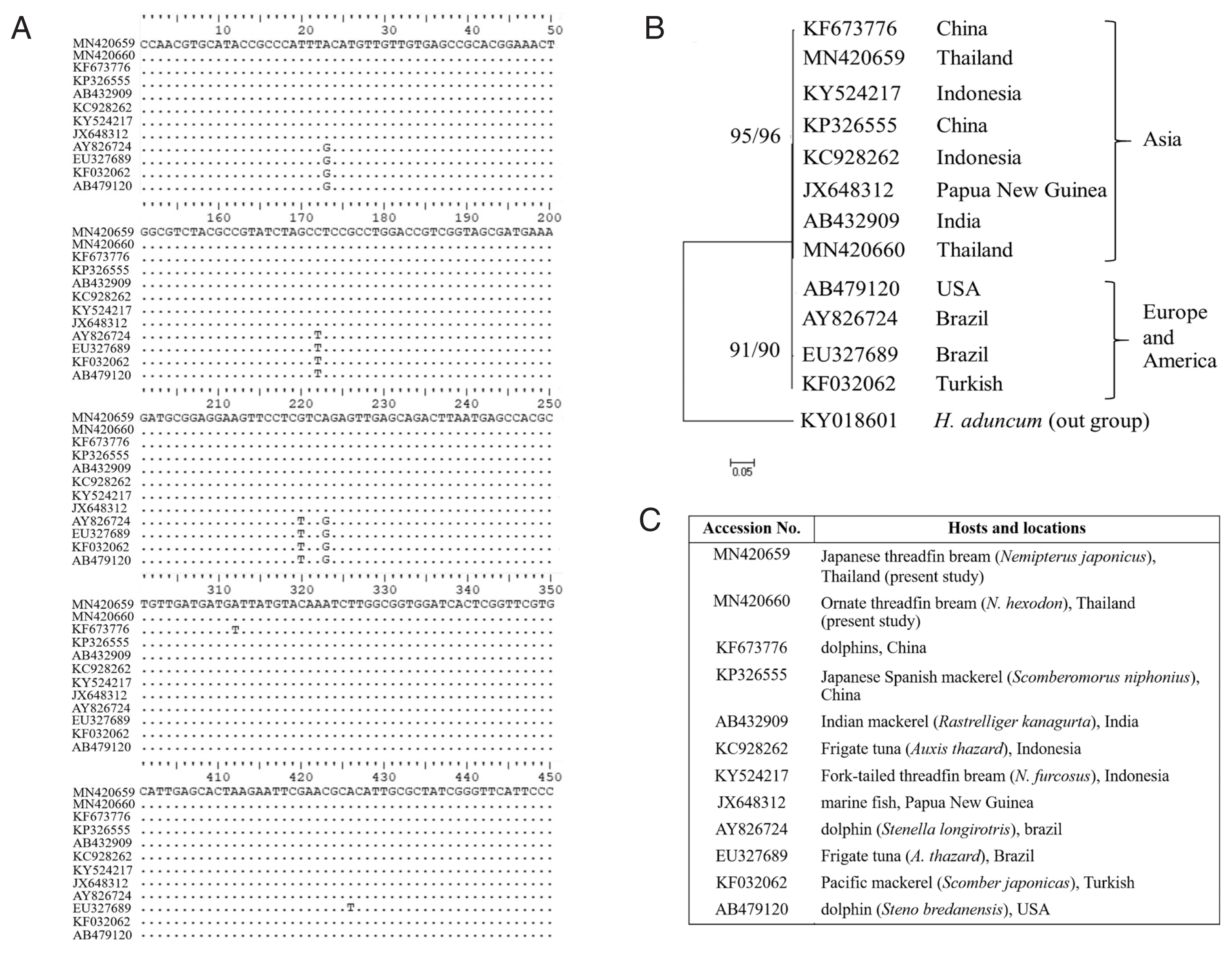
A. typica from Asia (China, India, Indonesia and Papua New Guinea) were separated from the group of
A. typica from Europe and America (
Fig. 7).
DISCUSSION
This is the first epidemiological study of L3 anisakid larvae in ornate threadfin bream (
N. hexodon) and Japanese threadfin bream (
N. japonicus) in Ching Mai province, northern Thailand. Here we have identified the species using both morphological and molecular analyses. The L3 larvae of
A. typica in the present study occurred at a high prevalence, intensity and abundance, the prevalence, intensity and abundance in
N. hexodon were 68.0%, 3.47, and 2.36, respectively, while in
N. japonicus the values were 60.0%, 4.17, and 2.50, respectively. The first study of the anisakid nematodes from marine fish in the Gulf of Thailand has been reported [
25]. They identified adult-stage anisakid nematodes in Perciformes fish, including
Johnius carouna and
Dendrophysa russelli, and identified the helminths as
A. simplex morphologically.
Based on morphological characteristics observed using both light and scanning electron microscopy, L3 anisakid larvae in the present study were morphologically similar to previous specimens studied [
19,
22,
26–
30]. For example, a distinct boring tooth and undeveloped lips at the anterior end of larva, the position of the excretory pore at the base of ventrolateral lips below the boring tooth and the rounded tail with a mucron were similar to those previously described [
31]. However, L3 larvae of
A. simplex and
A. typica are difficult to differentiate morphologically because they look quite similar, especially with respect to the morphology of the anterior portion of the larvae, which consists of a boring tooth and undeveloped lips, the oblique esophago-intestinal junction and the posterior part with a terminal mucron. The examination of morphometric data in
Table 2 revealed that
A. simplex is substantially larger than
A. typica. Further, the protruded mucrons observed using a scanning electron micrograph (
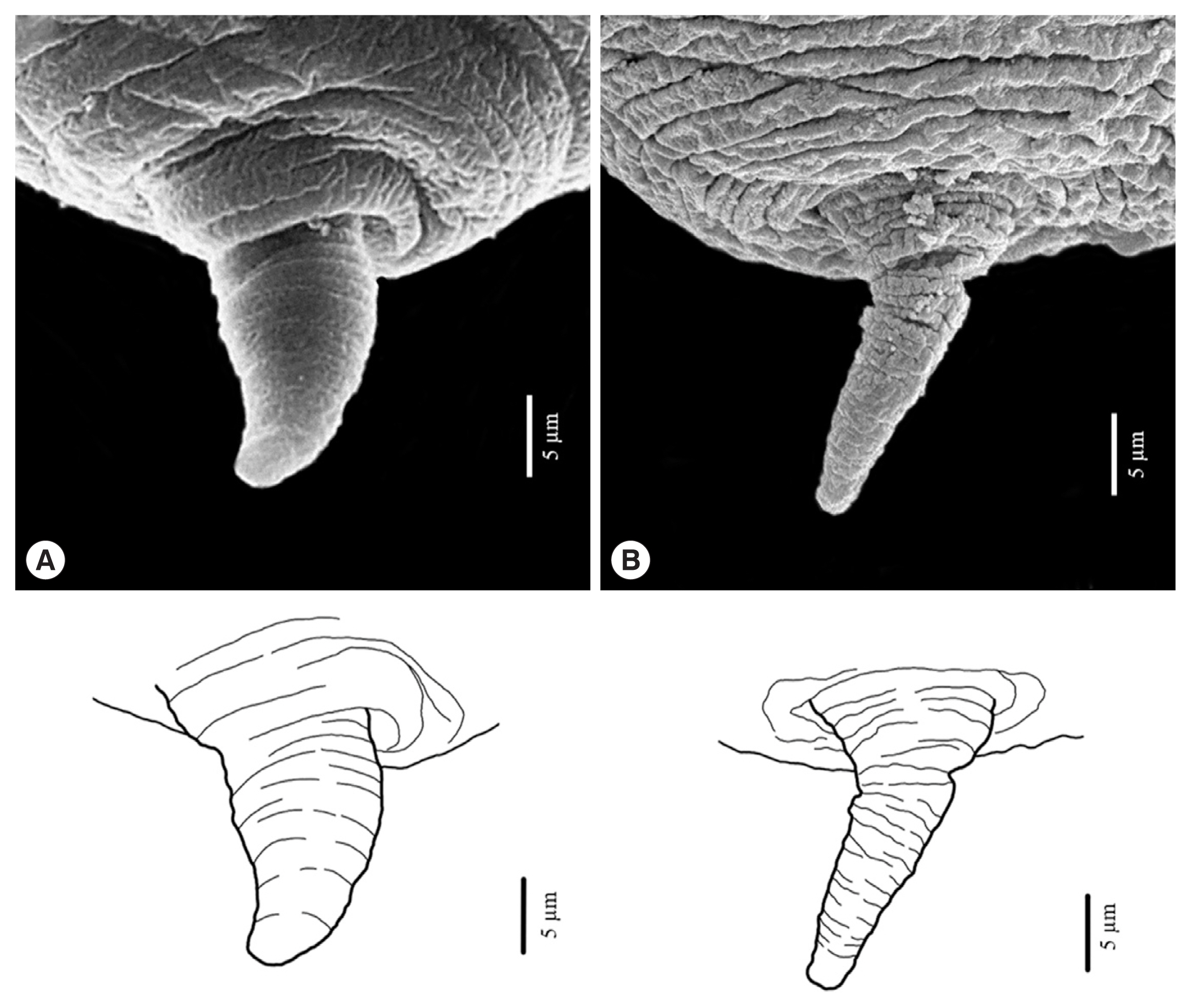
Fig. 5) showed
A. simplex from Ishii et al. [
24] was cone-shape, while
A. typica in the present study was cylindrical-shape, which seems narrower and longer than
A. simplex. Thus, the protruded mucron of L3 larvae should be able to be used to identify the larval species of anisakid larvae, but further study will be required to enhance the certainty of identification.
Molecular identification methods, such as PCR-amplification of genomic DNA within the ITS region of rDNA, is commonly used for identification of anisakid larvae in many countries and at least 6 species have been reported [
11–
17]. In Thailand, the molecular study of anisakid nematodes in marine fishes is still limited and no reports of anisakid nematodes from threadfin bream based on the ITS region of rDNA. So, our study is the first study of anisakid nematodes in threadfin bream from the Gulf of Thailand using the ITS region of rDNA. The previous study of the anisakid larvae in marine fish,
Priacanthus tayenus, from the Gulf of Thailand was identified as
A. typica based on mtDNA
cox2 with the genetic distance between 0.000–0.076 or 0–7.6% [
32]. Compared with the present study of the anisakid larvae in differences fish (
N. hexodon and
N. japonicus), we identified the larvae as
A. typica based on ITS region of rDNA with genetic distance=0.000 or 0% (
Table 3) and percent identity=100. Therefore, the genetic marker, ITS region of rDNA, in the present study is suitable for use to identify the species of the larvae. Moreover, according to the results of trees created using ML and NJ derived from ITS region of rDNA nucleotide sequences in the present study (
Fig. 7) showed that the group of
A. typica from marine animals in Asia were noticeably separated from groups of
A. typica from Europe and America. It seems that
A. typica from Asia, Europe and America exhibit diversity with respect to genetics and morphology.
It is well known that almost all reports of anisakiasis in humans have occurred by the accidental infection of
A. simplex and
A. pegreffii [
33,
34], but no report of anisakiasis in humans caused by
A. typica exists. However, a report has shown that this species can infect and cause severe tissue damage in the stomach of rats [
6]. Therefore,
A. typica may have a high impact on animals or even cause human infections.
Notes
-
The authors declare no conflict of interest related to this study.
ACKNOWLEDGMENTS
We would like to thank Dr. Preeyaporn Butboonchu and all members of the Applied Parasitology Research Laboratory, Department of Biology, Faculty of Science, Chiang Mai University. This research was partially supported by Chiang Mai University, Chiang Mai, Thailand, and the Institute for the Promotion of Teaching Science and Technology (IPST). Finally, we would like to thank
Proof-Reading-Service.com for assistance with the English language.
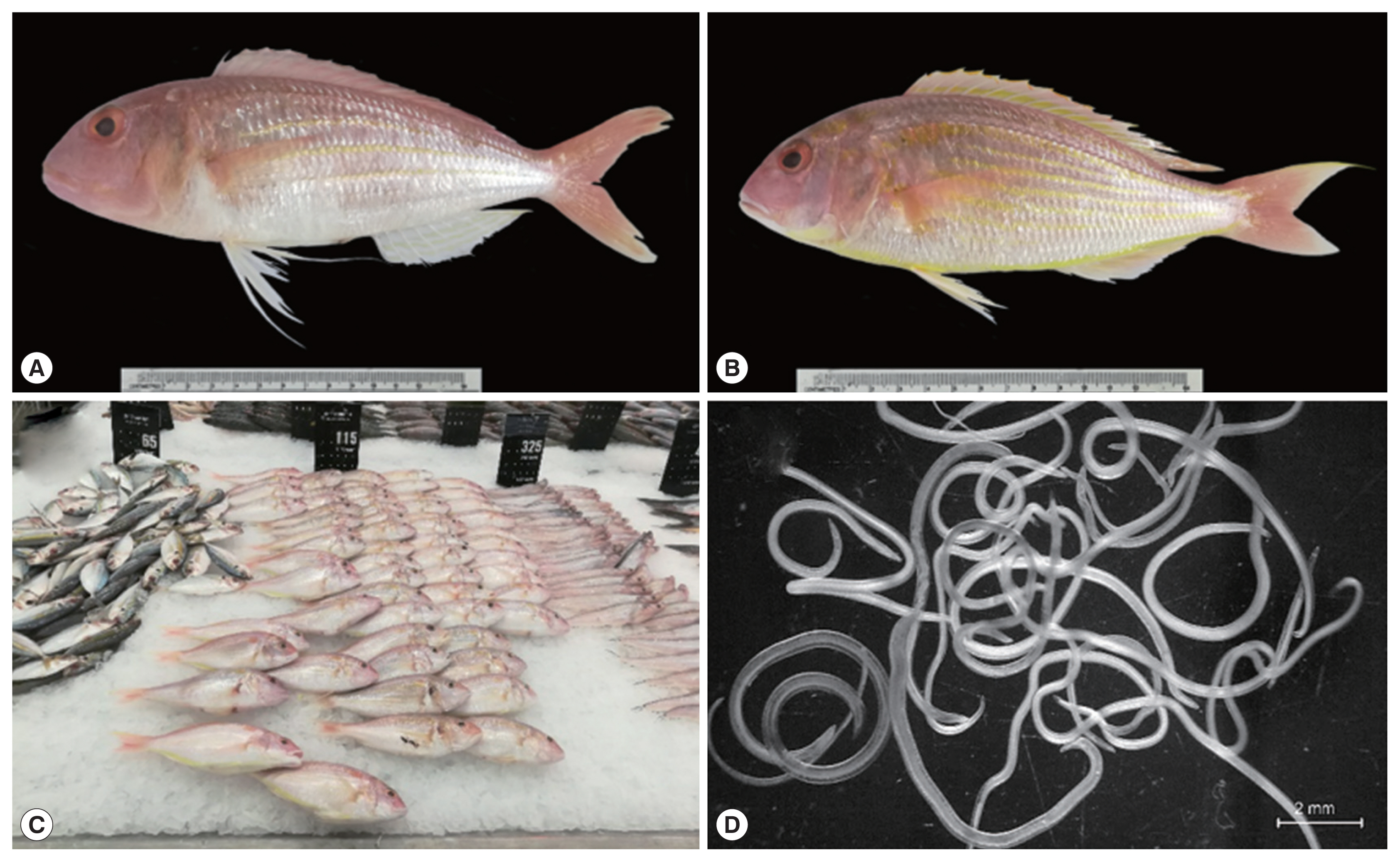
Fig. 1Ornate threadfin bream (N. hexodon) and Japanese threadfin bream (N. japonicus) (A and B, respectively) from the Gulf of Thailand purchased from a department store in Chiang Mai province. Fish sold in the department store in Chiang Mai province (C) and fresh specimens of L3 anisakid larvae viewed under a stereo microscope (D).
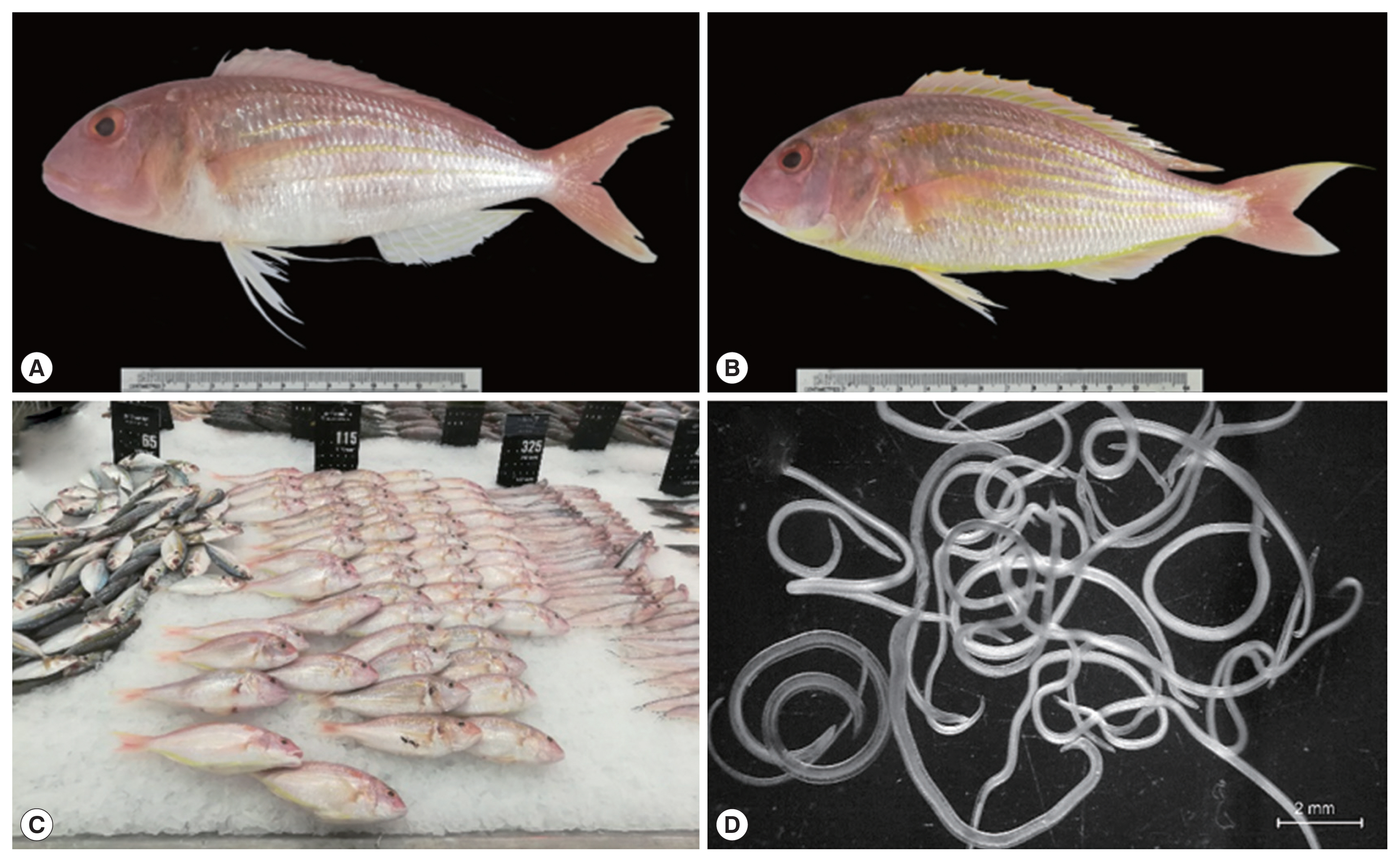
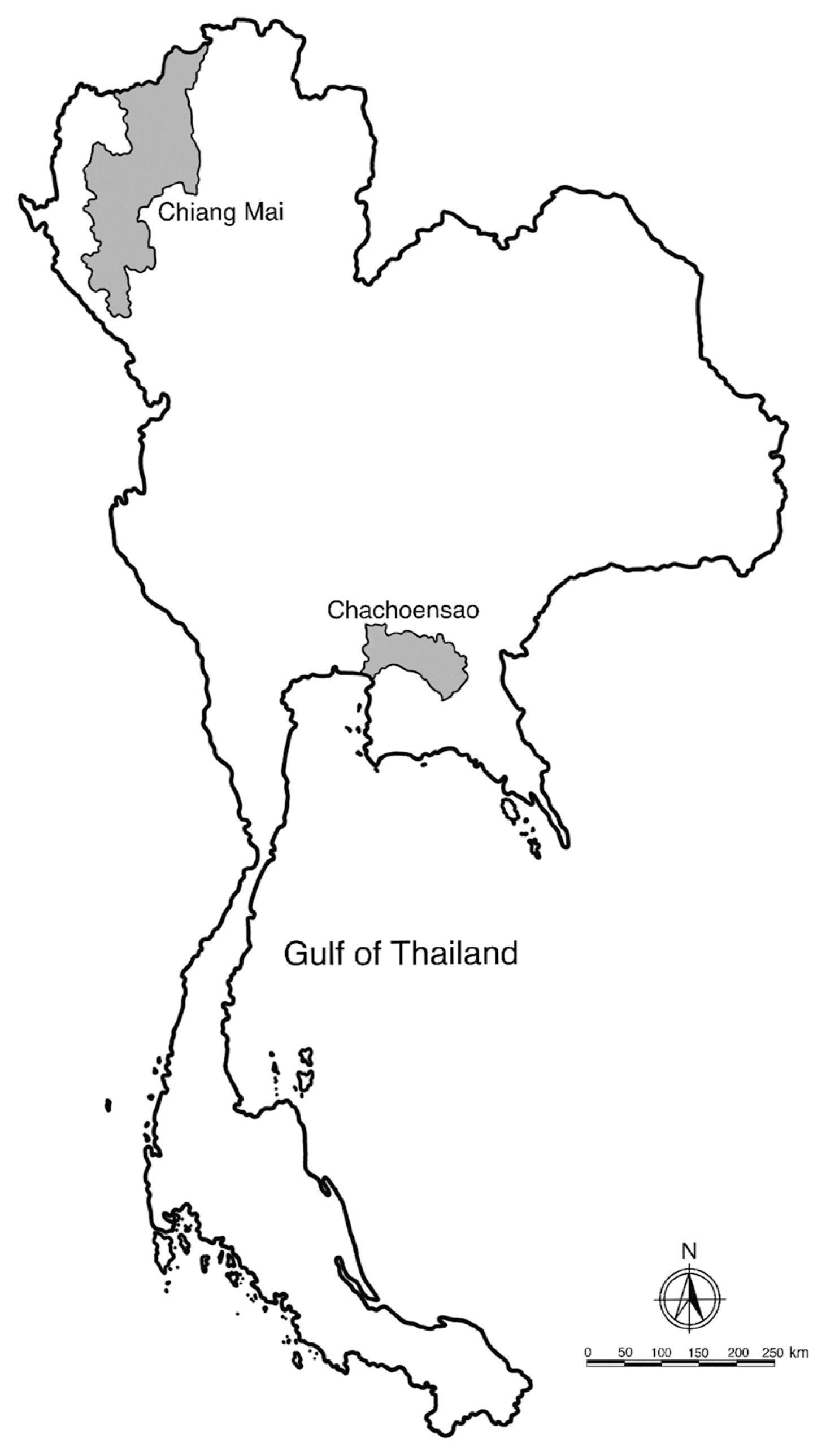
Fig. 2A map showing the location of Chachoengsao and Chiang Mai province, Thailand. Distance between the 2 locations is about 750 km.
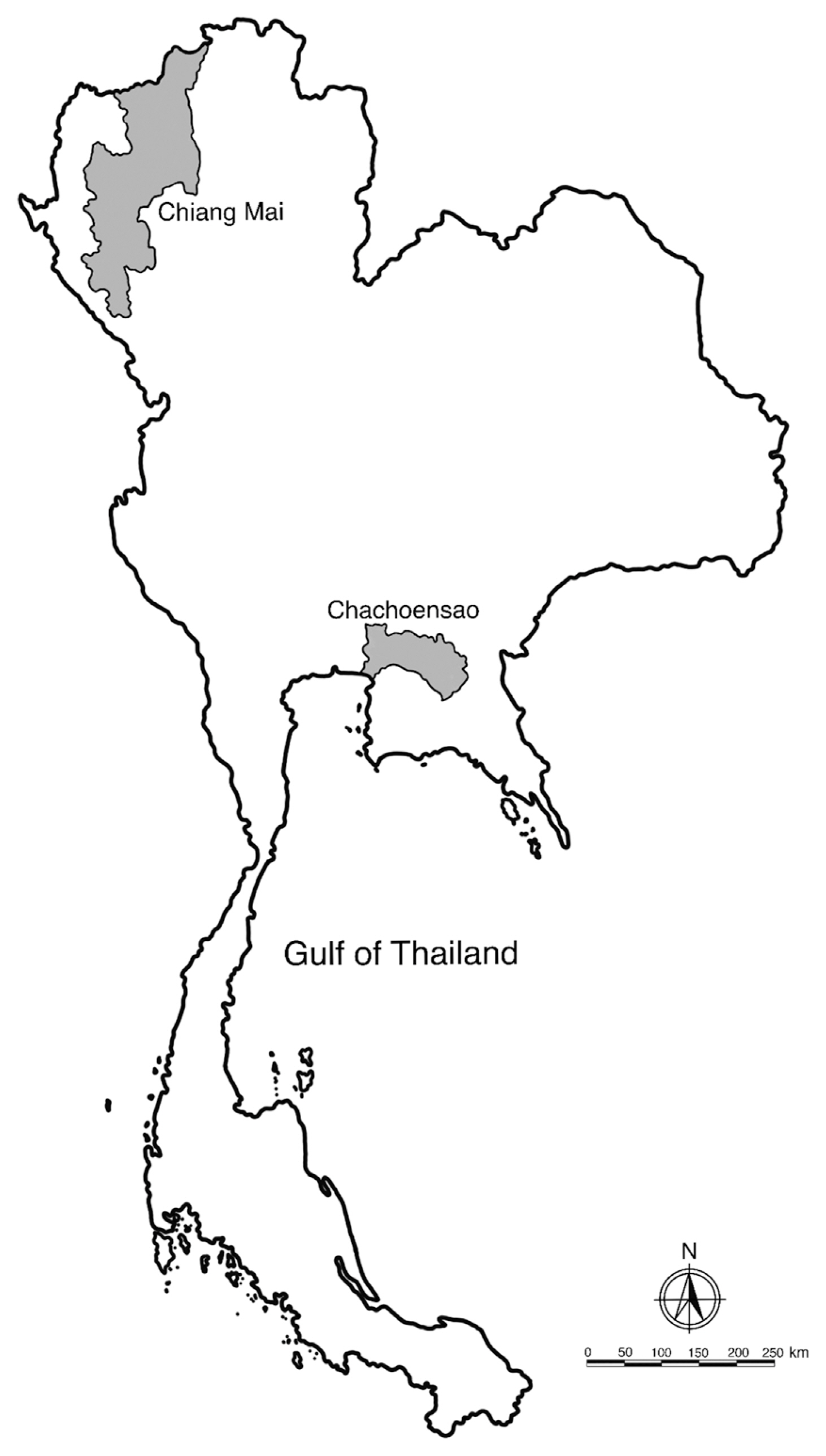
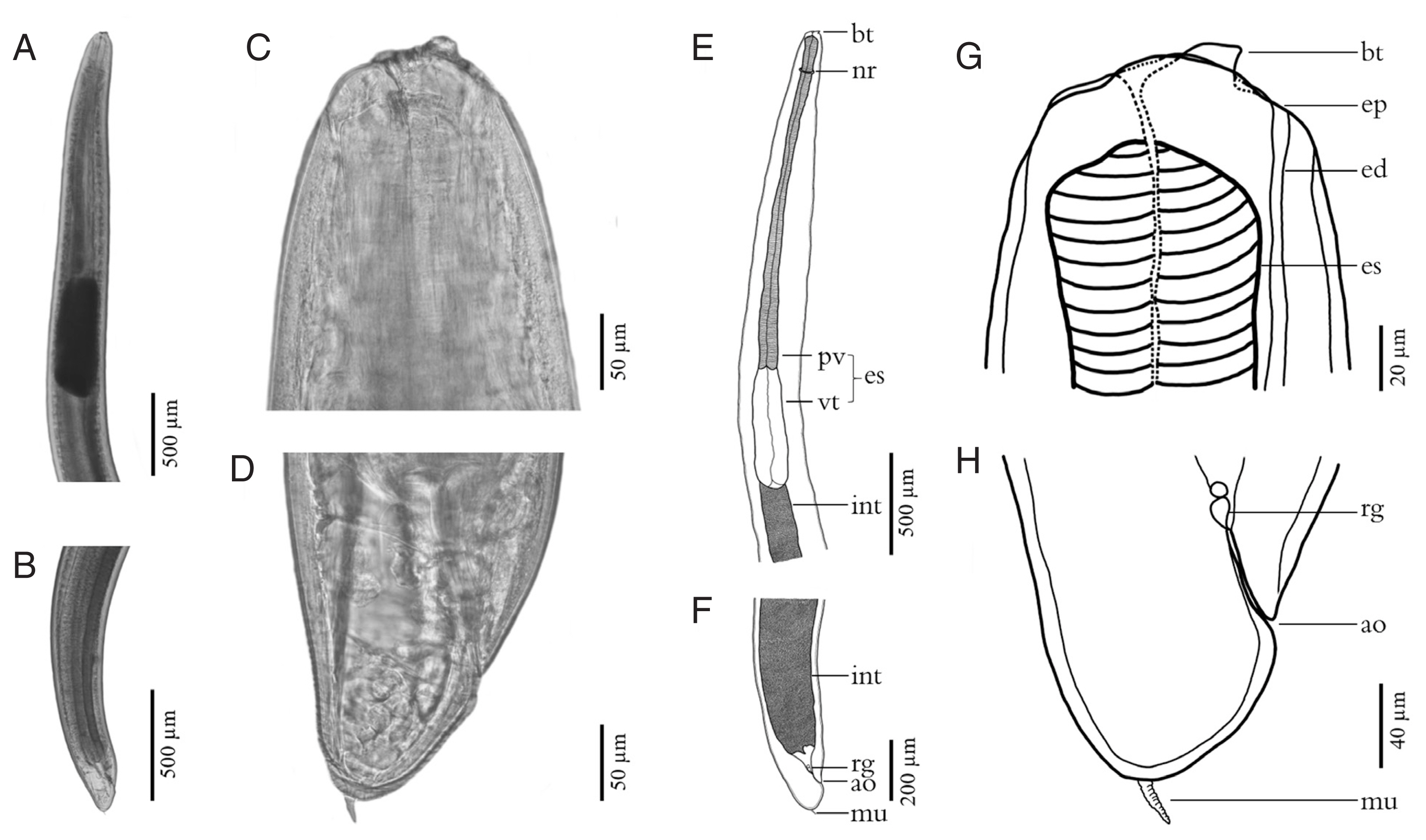
Fig. 3Fresh specimens of L3 A. typica from N. japonicus viewed under a light microscope are shown (A–D). Drawings on specimen from permanent slides (E–H) indicate the nerve ring, oblique esophago-intestinal junction (A, E) and tail (B, F), boring tooth and inconspicuous lips (C, G), rectal gland, anus and rounded tail with terminal mucron (D, H). ao, anus opening; bt, boring tooth; ed, excretory duct; ep, excretory pore; es, esophagus; int, intestine; mu, mucron; nr, nerve ring; pv, proventriculus; rg, rectal gland; vt, ventriculus.
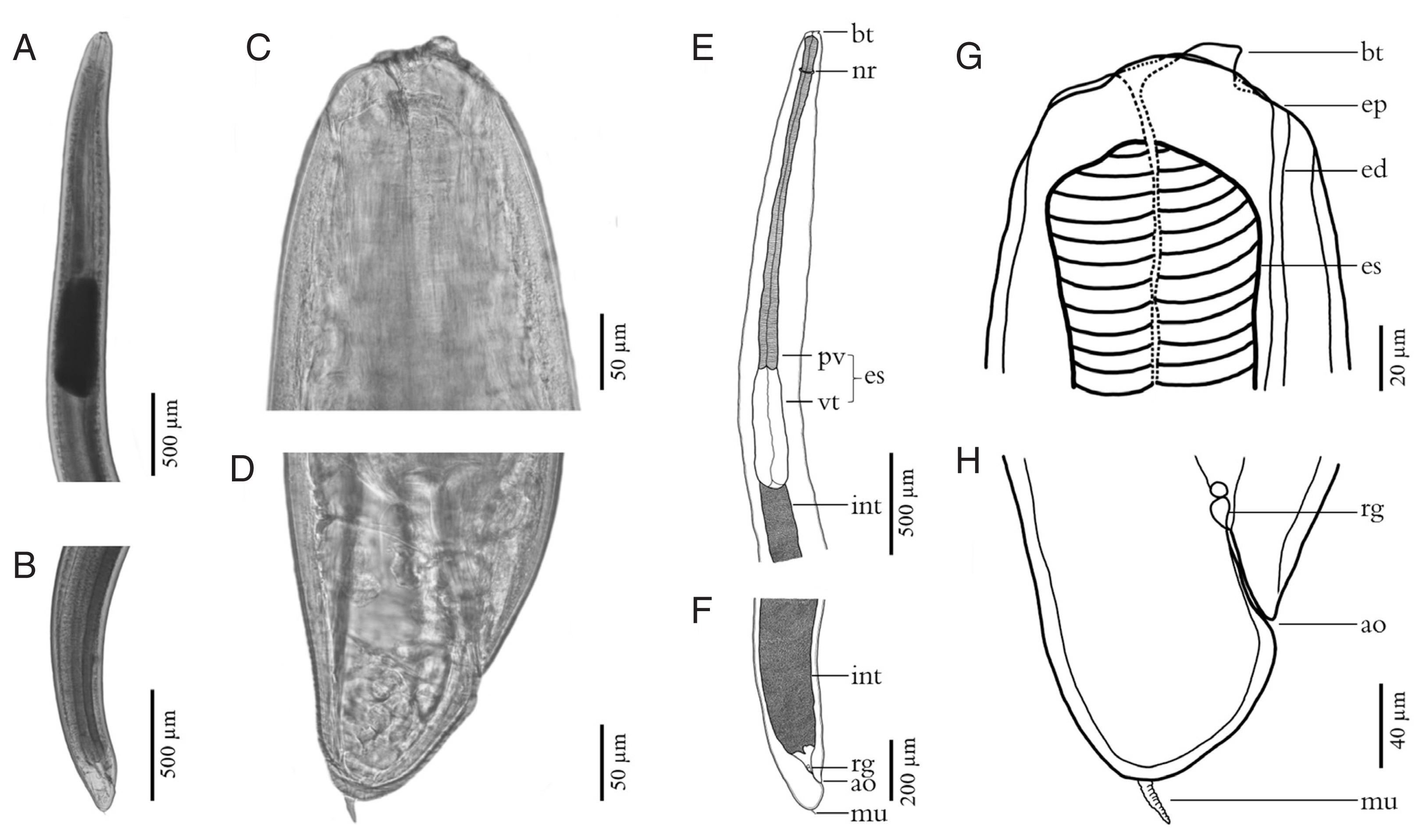
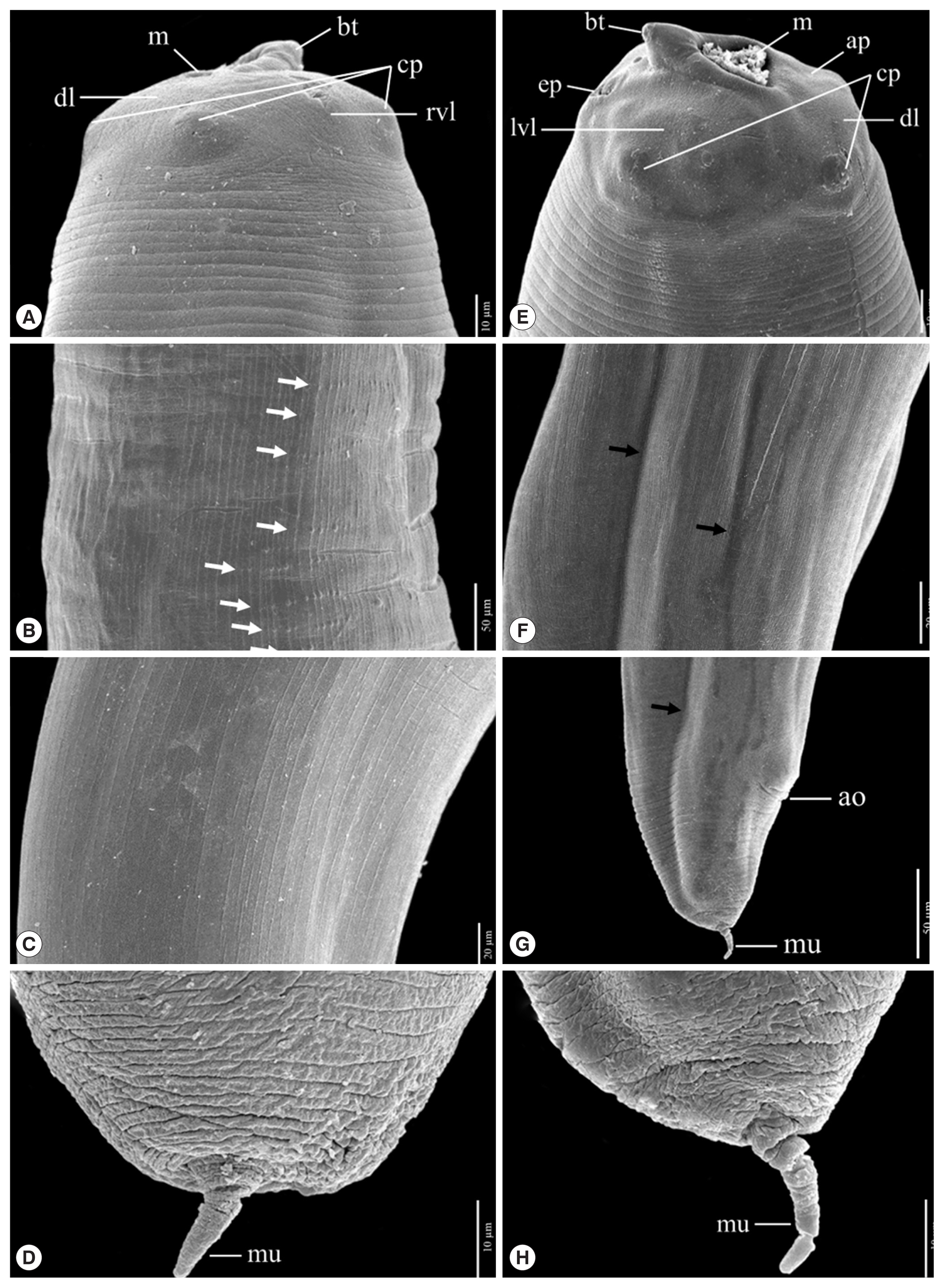
Fig. 4Scanning electron micrographs of L3 A. typica from N. hexodon (A–D) and N. japonicus (E–H) from the Gulf of Thailand. Mouth parts have been shown in A and E, cuticle structures in the midsection with longitudinal linear cuticular striations, small papillae (white arrow) and grooves (black arrow) (B, F). Cuticle structures of the posterior end with longitudinal linear cuticular striations (C), the posterior end including the anal opening and grooves (black arrow) (G) and posterior extremity with mucron (D, H) ao, anus opening; ap, amphid; bt, boring tooth; cp, cephalic papillae; dl, dorsal lip; ep, excretory pore; lvl, left ventrolateral lip; m, mouth; mu, mucron; rvl, right ventrolateral lip.
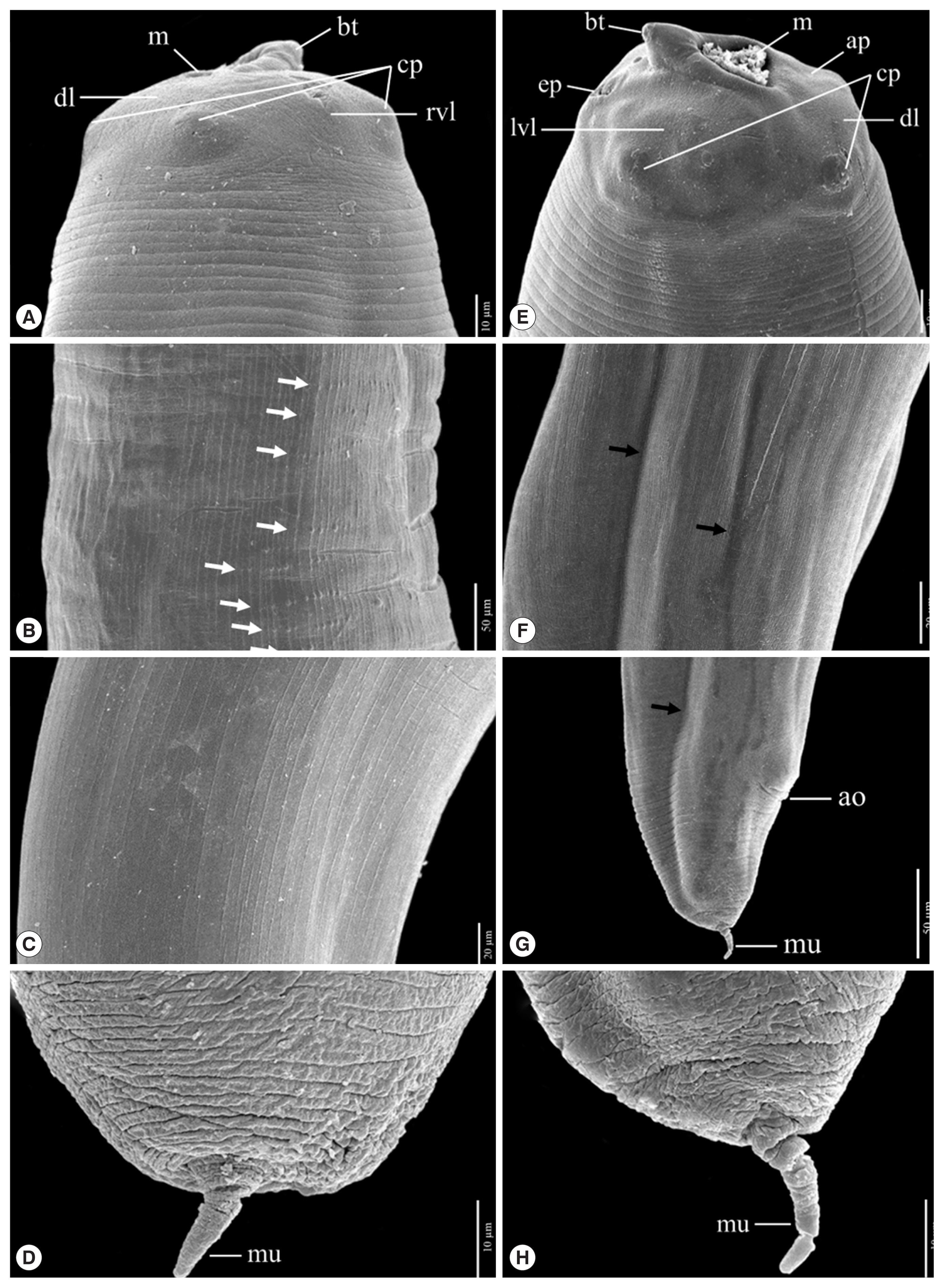
Fig. 5A comparison of the protruded mucron of L3 anisakid larvae using a scanning electron micrograph. Anisakis simplex from Ishii et al., 1989 has been shown in (A) and A. typica from the present study has been shown in (B).
Fig. 6Maximum likelihood (A) and neighbour-joining (B) phylogenetic trees for Anisakis spp. based on the ITS1-5.8S-ITS2 region of rDNA. Accession numbers are reported in the text and Hysterothylacium aduncum was used as an outgroup. Number on node is 1,000 bootstrap replications.
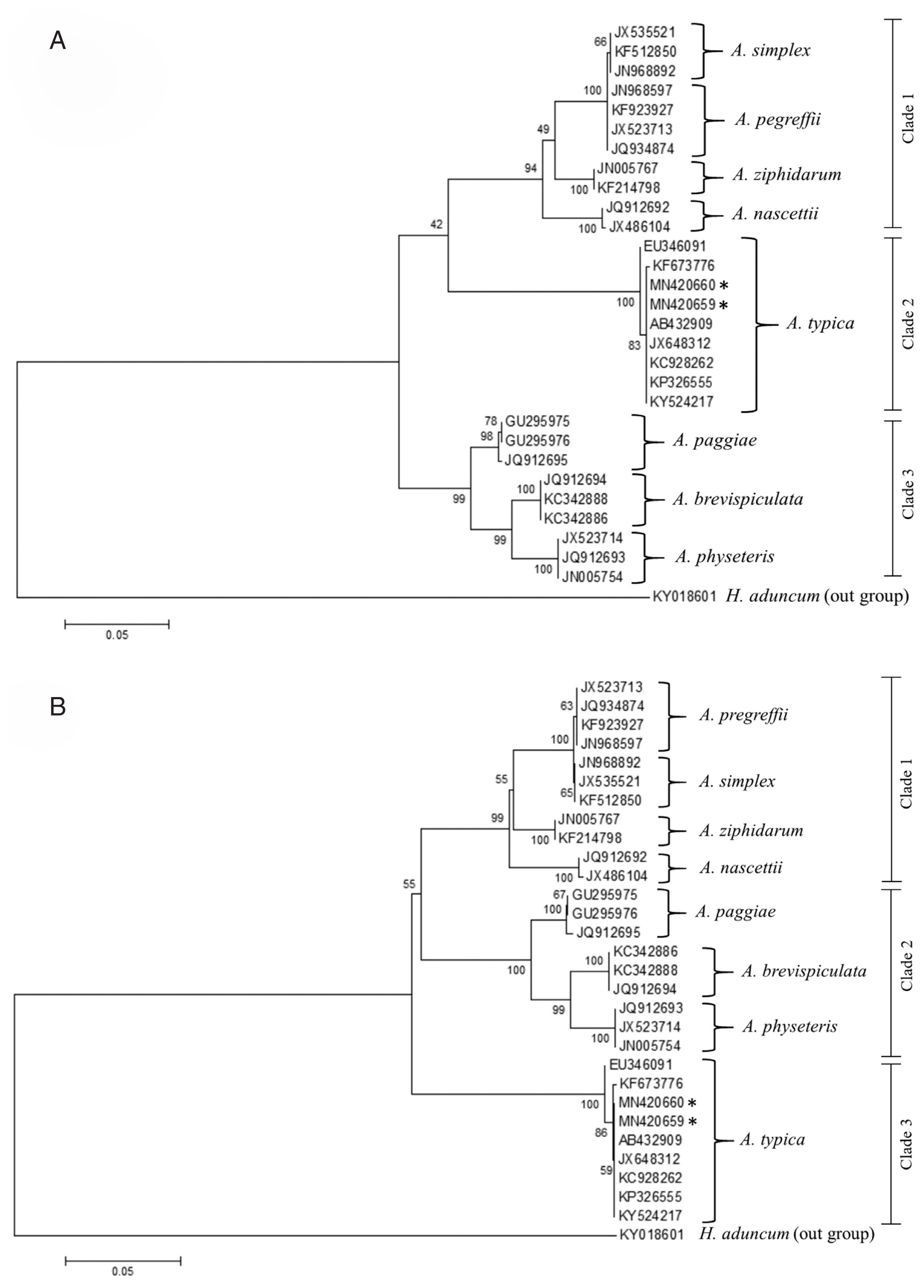
Fig. 7Comparisons of L3 A. typica ITS1-5.8S-ITS2 rDNA sequences in present study with others (A). Maximum likelihood and neighbour-joining phylogenetic tree (B) of A. typica based on ITS1-5.8S-ITS2 of rDNA with accession numbers, hosts and locations (C). Number of bootstrap replications was 1,000.
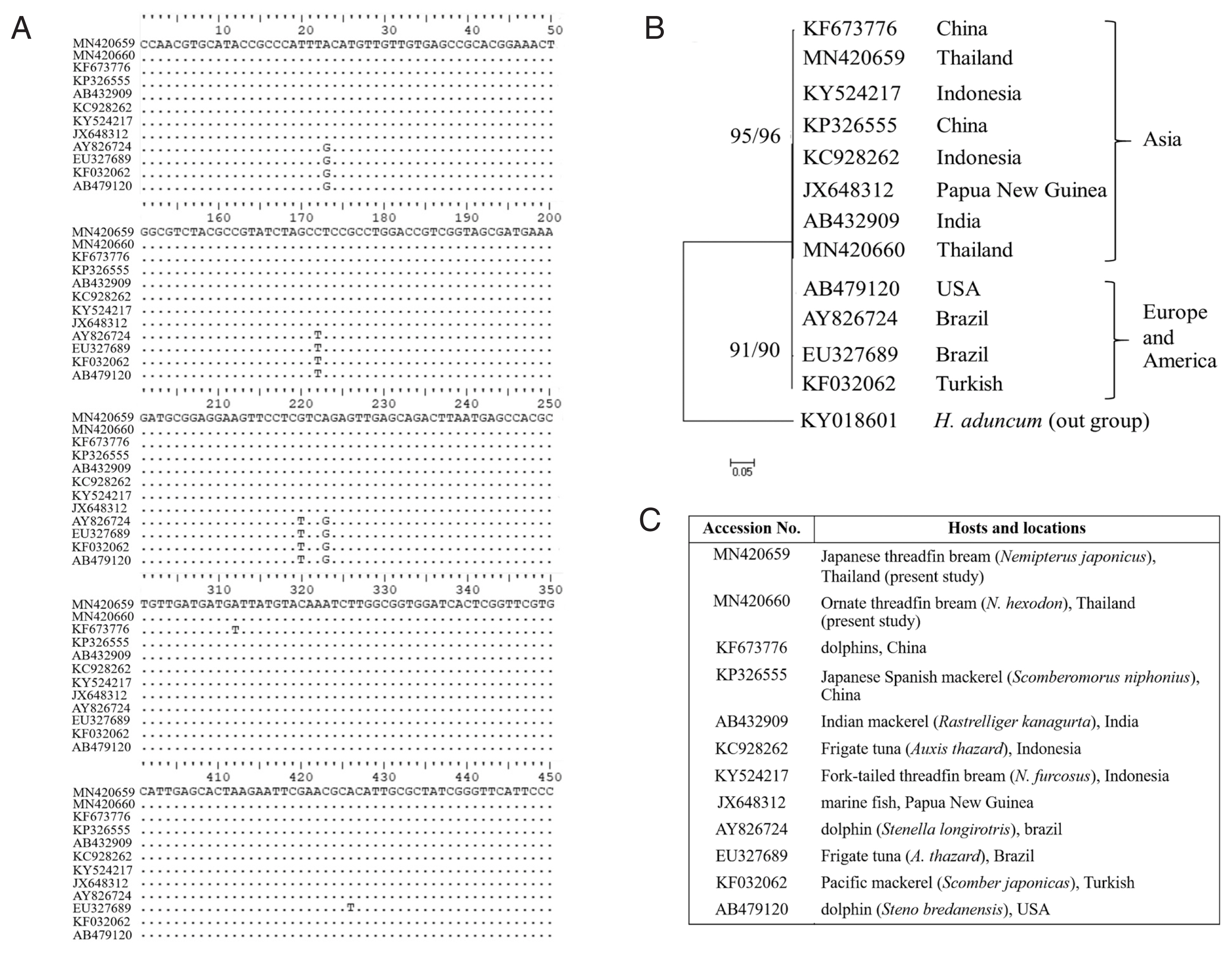
Table 1Average length and weight of sampled fish. Data regarding prevalence, intensity and sites infected with the L3 anisakid larvae have been given.
Table 1
|
Fish species |
Mean fish length (cm) |
Mean fish weight (g) |
Prevalence of anisakid larvae |
Intensity of anisakid larvae |
Abundance of anisakid larvae |
Infected sites (%) |
|
Body cavity |
Liver |
|
Ornate threadfin bream (Nemipterus hexodon) |
19.18 |
200.16 |
68% |
3.47 |
2.36 |
100.00 |
- |
|
Japanese threadfin bream (N. japonicus) |
18.42 |
162.63 |
60% |
4.17 |
2.50 |
99.20 |
0.80 |
Table 2Comparison of morphometric data obtained from L3 anisakid larvae (mm)
Table 2
|
Anisakis simplex
|
A. simplex
|
A. simplex
|
A. simplex
|
A. typica
|
A. typica
|
A. typica
|
A. typica
|
|
Hosts |
Oncorhynchus keta
|
Tribolodon hakanensis
|
Merluccius merluccius lessepsianus
|
O. keta
|
Pseudopercis numida
|
-
|
Trichiurus lepturus
|
Nemipterus hexodon & N. japonicus
|
|
Body length |
19.73–28.41 |
32.96–35.66 |
14.1–25.6 |
12.75–29.94 |
- |
- |
15.34–22.43 |
12.31–18.00 |
|
Body width |
0.42–0.68 |
0.6 |
0.48–0.62 |
0.45–0.75 |
- |
- |
0.35–0.60 |
0.34–0.45 |
|
Proventriculus length |
1.03–2.59 |
2.58 |
1.18–2.68 |
1.18–2.58 |
1.52 |
1.32–1.57 |
1.10–1.81 |
1.36–1.88 |
|
Proventriculus width |
- |
- |
- |
- |
- |
- |
- |
0.08–0.12 |
|
Ventriculus length |
0.83–1.56 |
1.16–1.52 |
0.71–0.92 |
0.90–1.50 |
0.62 |
0.48–0.88 |
0.50–0.76 |
0.55–0.94 |
|
Ventriculus width |
0.14–0.35 |
0.20–0.25 |
- |
0.13–0.31 |
- |
- |
- |
0.10–0.20 |
|
Mucron length |
0.01–0.03 |
0.015 |
0.019–0.032 |
0.02–0.03 |
- |
- |
- |
0.01–0.02 |
|
References |
[18] |
[23] |
[24] |
[25] |
[14] |
[26] |
[27] |
Present study |
Table 3Genetic distance of Anisakis spp. based on ITS1-5.8S-ITS2 of rDNA
Table 3
|
Taxa |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
Anisakis typicaa
|
- |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. typicab
|
0.000 |
- |
0.000 |
0.000 |
0.000 |
0.000 |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. typica (AB432909) |
0.000 |
0.000 |
- |
0.000 |
0.000 |
0.000 |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. typica (HF911524) |
0.000 |
0.000 |
0.000 |
- |
0.000 |
0.000 |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. typica (JX523715) |
0.000 |
0.000 |
0.000 |
0.000 |
- |
0.000 |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. typica (KC928262) |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
- |
0.021 |
0.021 |
0.021 |
0.022 |
0.022 |
0.023 |
0.023 |
0.202 |
|
A. ziphidarum
|
0.159 |
0.159 |
0.159 |
0.159 |
0.159 |
0.159 |
- |
0.016 |
0.018 |
0.019 |
0.009 |
0.009 |
0.009 |
0.205 |
|
A. paggiae
|
0.159 |
0.159 |
0.159 |
0.159 |
0.159 |
0.159 |
0.124 |
- |
0.009 |
0.008 |
0.017 |
0.017 |
0.018 |
0.202 |
|
A. physeteris
|
0.164 |
0.164 |
0.164 |
0.164 |
0.164 |
0.164 |
0.140 |
0.054 |
- |
0.007 |
0.019 |
0.019 |
0.021 |
0.208 |
|
A. brevispiculata
|
0.172 |
0.172 |
0.172 |
0.172 |
0.172 |
0.172 |
0.143 |
0.047 |
0.035 |
- |
0.019 |
0.019 |
0.021 |
0.225 |
|
A. simplex
|
0.175 |
0.175 |
0.175 |
0.175 |
0.175 |
0.175 |
0.053 |
0.127 |
0.145 |
0.144 |
- |
0.001 |
0.011 |
0.205 |
|
A. pegreffii
|
0.177 |
0.177 |
0.177 |
0.177 |
0.177 |
0.177 |
0.052 |
0.125 |
0.143 |
0.146 |
0.001 |
- |
0.011 |
0.204 |
|
A. nascettii
|
0.182 |
0.182 |
0.182 |
0.182 |
0.182 |
0.182 |
0.053 |
0.141 |
0.162 |
0.160 |
0.070 |
0.068 |
- |
0.243 |
|
H. aduncumc
|
0.910 |
0.910 |
0.910 |
0.910 |
0.910 |
0.910 |
0.916 |
0.925 |
0.928 |
0.994 |
0.916 |
0.912 |
1.033 |
- |
References
- 1. Stobutzki IC, Silvestre GT, Talib AA, Krongprom A, Supongpan M, Khemakorn P, Armada N, Garces LR. Decline of demersal coastal fisheries resources in three developing Asian countries. Fish Res 2006;78:130-142.
- 2. Valls A, Pascual CY, Esteban MM.
Anisakis allergy: an update. Rev Fr Allergol 2005;45:108-113.
- 3. Nieuwenhuizen NE, Lopata AL.
Anisakis – A food-borne parasite that triggers allergic host defences. Int J Parasitol 2013;43:1047-1057.
- 4. Mladineo I, Popović M, Hofman ID, Poljak V. A case report of Anisakis pegreffii (Nematoda, Anisakidae) identified from archival paraffin sections of a Croatian patient. BMC Infect Dis 2016;16:1-5.
- 5. Jelena I, Milan ZB, Marija B, Natasa K, Marija D, Radmila M, Jelena J, Branislav B.
Anisakis allergy in human. Trends Food Sci Tech 2017;59:25-29.
- 6. Soewarlan LC, Suprayitno E, Hardoko , Nursyam H. A model of rat’s stomach orally infected by live Anisakis typica larvae: a histopatological study. Int J Pharm Tech Res 2015;8:221-229.
- 7. Nawa Y, Hatz C, Blum J. Sushi delights and parasites: The risk of fishborne and foodborne parasitic zoonoses in Asia. J Travel Med 2005;41:1297-1303.
- 8. Carrascosa MF, Mones JC, Caviedes JS, Román JG. A man with unsuspected marine eosinophilic gastritis. Lancet Infect Dis 2015;15:248.
- 9. Chen WC. A case of human infection with Anisakis simplex in Taiwan. Gastrointest Endosc 2015;82:757-758.
- 10. Nogami Y, Nishimura YF, Banno K, Suzuki A, Susumu N, Hibi T, Murakami K, Yamada T, Sugiyama H, Morishima Y, Aoki D. Anisakiasis mimics cancer recurrence: two cases of extragastrointestinal anisakiasis suspected to be recurrence of gynecological cancer on PET-CT and molecular biological investigation. BMC Med Imaging 2016;16:31.
- 11. Zhu X, Gasser RB, Podolska M, Chilton NB. Characterisation of anisakid nematodes with zoonotic potential by nuclear ribosomal DNA sequences. Int J Parasitol 1998;28:1911-1921.
- 12. Palm HW, Damriyasa IM, Linda , Oka IBM. Molecular genotyping of Anisakis Dujardin, 1845 (Nematoda: Ascaridoidea: Anisakidae) larvae from marine fish of Balinese and Javanese waters, Indonesia. Helminthologia 2008;45:3-12.
- 13. Iniguez AM, Santos CP, Vicente ACP. Genetic characterization of Anisakis typica and Anisakis physeteris from marine mammals and fish from the Atlantic Ocean off Brazil. Vet Parasitol 2009;165:350-356.
- 14. Umehara A, Kawakami Y, Ooi HK, Uchida A, Ohmae H, Sugiyama H. Molecular identification of Anisakis type I larvae isolated from hairtail fish off the coasts of Taiwan and Japan. Int J Food Microbiol 2010;143:161-165.
- 15. Iniguez AM, Carvalho VL, Motta MRA, Pinheiro DCSN, Vicente ACP. Genetic analysis of Anisakis typica (Nematoda: Anisakidae) from cetaceans of the northeast coast of Brazil: new data on its definitive hosts. Vet Parasitol 2011;178:293-299.
- 16. Kong Q, Fan L, Zhang J, Akao N, Dong K, Lou D, Ding J, Tong Q, Zheng B, Chen R, Ohta N, Lu S. Molecular identification of Anisakis and Hysterothylacium larvae in marine fishes from the East China Sea and the Pacific coast of central Japan. Int J Food Microbiol 2015;199:1-7.
- 17. Chen HY, Shih HH. Occurrence and prevalence of fish-borne Anisakis larvae in the spotted mackerel Scomber australasicus from Taiwanese waters. Acta Trop 2015;145:61-67.
- 18. Russell BC. A review of the threadfin breams of the genus Nemipterus (Nemipteridae) from Japan and Taiwan, with description of a new species. Jpn J Ichthyol 1993;39:295-310.
- 19. Pantoja CS, Borges JN, Santos CP, Luque JL. Molecular and morphological characterization of anisakid nematode larvae from the sandperches Pseudopercis numida and Pinguipes brasilianus (Perciformes: Pinguipedidae) off Brazil. J Parasitol 2015;101:492-499.
- 20. Yamaguti S. Systema Helminthum. 3:Nematodes. 1st ed.. New York, USA. Interscience Publishers Inc; 1961, pp 583-586.
- 21. Murata R, Suzuki J, Kai A. Morphological and molecular characterization of Anisakis larvae (Nematoda: Anisakidae) in Beryx splendens from Japanese waters. Parasitol Int 2011;60:193-198.
- 22. Setyobudi E, Jeon CH, Lee C, Seong KB, Kim JH. Occurrence and identification of Anisakis spp. (Nematoda: Anisakidae) isolated from chum salmon (Oncorhynchus keta) in Korea. Parasitol Res 2011;108:585-592.
- 23. Koinari M, Karl S, Elliot A, Ryan U, Lymbery AJ. Identification of Anisakis species (Nematoda: Anisakidae) in marine fish hosts from Papua New Guinea. Vet Parasitol 2013;193:126-133.
- 24. Ishii Y, Fujino T, Weerasooriya MV. Morphology of anisakine larvae. In Ishikura H, Namiki M eds, Gastric Anisakiasis in Japan. Tokyo, Japan. Springer; 1989, pp 19-29.
- 25. Nuchjangreed C, Hamzah Z, Suntornthiticharoen P, Nuntawarasilp PS. Anisakids in marine fish from the coast of Chonburi province, Thailand. Southeast Asian J Trop Med Public Health 2006;37:35-39.
- 26. Moravec F, Nagasawa K, Urawa S. Some fish nematodes from fresh waters in Hokkaido, Japan. Folia Parasitol 1985;32:305-316.
- 27. Rahma YA, Gaber RA, Ahmed AK. First record of Anisakis simplex third-stage larvae (Nematoda, Anisakidae) in European hake Merluccius merluccius lessepsianus in Egyptian water. J Parasitol Res 2016;2016:1-8.
- 28. Quiazon KMA, Yoshinaga T, Ogawa K, Yukami R. Morphological differences between larvae and in vitro-cultured adults of Anisakis simplex (sensu stricto) and Anisakis pegreffii (Nematoda: Anisakidae). Parasitol Int 2008;7:483-489.
- 29. Jabbar A, Asnoussi A, Norbury LJ, Eisenbarth A, Shamsi S, Gasser RB, Lopata AL, Beveridge I. Larval anisakid nematodes in teleost fishes from Lizard Island, northern Great Barrier Reef, Australia. Mar Freshwater Res 2012;63:1283-1299.
- 30. Borges JN, Cunha LFG, Santos HLC, Neto CM, Santos CP. Morphological and molecular diagnosis of anisakid nematode larvae from cutlassfish (Trichiurus lepturus) off the coast of Rio de Janeiro, Brazil. PLoS One 2012;7:1-14.
- 31. Serrano TD, Pelegrini LS, Santiago ET, Prado FD, Foresti FP, Azevedo RK, Abdallah VD. Molecular identification and morphological characterization of Anisakis spp. L3 larvae (Nematoda: Anisakidae) in Scomber colias Gmelin, 1789 (Perciformes: Scombridae) from northern Argentina. Neotrop Helminthol 2017;11:25-36.
- 32. Eamsobhana P, Yong HS, Song SL, Tungtrongchitr A, Roongruangchai K. Genetic differentiation of Anisakis species (Nematoda: Anisakidae) in marine fish Priacanthus tayenus from Gulf of Thailand. Trop Biomed 2018;35:669-677.
- 33. Mattiucci S, Paoletti M, Borrini F, Palumbo M, Palmieri RM, Gomes V, Casati A, Nascetti G. First molecular identification of the zoonotic parasite Anisakis pegreffii (Nematoda: Anisakidae) in a paraffin-embedded granuloma taken from a case of human intestinal anisakiasis in Italy. BMC Infect Dis 2011;11:82.
- 34. Arizono N, Yamada M, Tegoshi T, Yoshikawa M.
Anisakis simplex sensu stricto and Anisakis pegreffii: biological characteristics and pathogenetic potential in human anisakiasis. Foodborne Pathog Dis 2012;9:517-521.