Molecular Identification of Taenia hydatigena from Sheep in Khartoum, Sudan
Article information
Abstract
The cestode Taenia hydatigena uses canids, primarily dogs, as definitive hosts, while the metacestode larval stage cysticercus infects a range of intermediate hosts, including domestic animals such as goats, sheep, and pigs. Cysticercosis due to T. hydatigena has large veterinary and economic drawbacks. Like other taeniids, e.g., Echinococcus, intraspecific variation is found among the members of the genus Taenia. In Africa, few studies are available on the epidemiology and distribution of T. hydatigena, and even fewer studies are available on its genetic variation. In this study, we molecularly identified 11 cysticerci from sheep in Sudan and demonstrated the genetic variation based on the NADH dehydrogenase subunit 1 (nad1) and cytochrome c oxidase subunit 1 (cox1) mitochondrial genes. The isolates were correctly identified as T. hydatigena with more than 99% similarity to those in the GenBank database. Low diversity indices and insignificant neutrality indices were observed, with 3 and 2 haplotypes for the nad1 and cox1 genes, respectively. The results suggest the presence of unique T. hydatigena haplotypes in Sudan, as haplotypes with 100% similarity were not found in the GenBank database. With few available studies on the genetic variation of T. hydatigena in Africa, this report represents the first insights into the genetic variation of T. hydatigena in Sudan and constitutes useful data.
The cestode Taenia hydatigena uses canids, primarily dogs, as definitive hosts, while the metacestode larval stage infects a range of intermediate hosts, including goats, sheep, and pigs. Cysticercosis due to T. hydatigena is of veterinary and economic importance, especially in small ruminants, where it causes production loss through clinical disease or damage to infected organs [1–3].
In Africa, a few studies are available on the epidemiology and distribution of T. hydatigena [4]. Prevalence reports, where available, have demonstrated high infection rates in sheep and goats and low infection rates in pigs [4–8]. In Sudan, data on the prevalence of T. hydatigena in livestock are scarce. However, a study conducted by El Badawi et al. [9] reported a high prevalence of T. hydatigena in sheep (32.4%) and goats (29.0%) and emphasized its economic importance and implications for livestock production.
Genetic variation has been well studied in most cestodes species, e.g., Echinococcus granulosus [10,11], and correlates with parasite morphology, host infectivity, drug and vaccine development, and ultimately control. Similarly, mitochondrial genome studies of Taenia spp. have also confirmed the existence of genetic variation within species [13,14], and in some cases, such variation has been suggested to correlate with pathological differences in different hosts [15,16]. The molecular epidemiology and genetic variation of T. hydatigena in Africa are underinvestigated; however, a few studies in countries such as Nigeria, Egypt and Tanzania have demonstrated nucleotide variation between isolates from sheep, goats, and pigs [4,17,18]. Meanwhile, studies describing the genetic population structure of T. hydatigena in Sudan and how they differ from other geographical regions, as well as the epidemiological significance, are lacking. Therefore, the aim of this study was to molecularly identify T. hydatigena of Sudanese origin and to investigate possible genetic variation based on the NADH dehydrogenase subunit 1 (nad1) and cytochrome c oxidase subunit 1 (cox1) mitochondrial genes.
Thirty cyst samples (2–5 cm in size) from liver were harvested from slaughtered sheep in various homes in the Western Nile (Al-Hajyosif and Al Gouz)- Khartoum, Sudan, during a meat/slaughtering inspection for the Addha festival (September 2017). A total of 1,500 sheep were examined during the inspection. However, the slaughtered sheep were said to have originated from North Kordofan State and the west White Nile State of Sudan (Fig. 1). DNA was extracted from a portion of each cysticercus using the phenol-chloroform extraction method. Extracted genomic DNA samples were stored at −20°C until use. PCR amplification of the mitochondrial nad1 and cox1 genes was performed using the following primer pairs specific for cestode parasites: forward 5′CARTTTCGTAAGGGBCCWAAWAAGGT and reverse 5′-CCAATTTCYTGAAGTTAACAGCATCA [19]; and a newly designed forward (5′-AGTCCTGATGCTTTTGGGTTCTATGGA-3′) and a previously reported reverse primer (5′-AAGCATGATGCAAAAGGCAAATAAACC-3′) [20], for the nad1 and cox1 genes, respectively, with expected fragment sizes of 871 and 939 bp, respectively. PCR was conducted in a 25 μl reaction mixture containing 12.5 μl Premix Ex TaqTM version 2.0 (Takara Bio, Japan), 10 pmol of each primer, 0.5 μl of genomic DNA extract (20–200 ng), and RNAse free water up to the final concentration of 25 μl. The reaction was performed under the following conditions: initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec and elongation at 72°C for 60 sec, and a final extension at 72°C for 10 min. Then, 5 μl of the PCR products were analyzed in a 1.5% (w/v) agarose gel stained with GelRedTM while the remainder was sequenced in an ABI3730Xl DNA Analyser (Beijing Tsingke Biotechnology Co., Beijing, PR China).
DNA sequences were viewed and manually corrected for any misread nucleotide and aligned using BioEdit software [21]. The identity of each isolate was confirmed in a BLAST search in the GenBank database using the NCBI BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The diversity (nucleotide and haplotype diversity) and neutrality (Fu’s Fs and Tajima’s D) indices were estimated in DnaSP v.6 [22]. Bayesian phylogeny was inferred based on the nad1 and cox1 dataset using MrBayes v.3.1.2. Markov chain Monte Carlo (MCMC) sampling was used to assess the posterior distribution of the parameters with a chain length of 2,000,000 states, and 10% was discarded as burn-in. Parameters were logged every 1,000 states. TreeView v.1.6.6. (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to draw/display trees.
Of the 30 cyst samples, successful amplification and sequencing of the nad1 and cox1 genes identified 11 cysts as T. hydatigena. No amplification was observed for the other 19 cysts, even after the process was repeated. The useable nucleotide sequences after correction and editing were 764 and 735 bp for nad1 and cox1, respectively. These sequences were more than 99% similar to those in the GenBank repository. The Sudanese T. hydatigena isolates showed the highest similarity (99.73–99.87%) to the Chinese and Nigerian isolates according to the BLAST query. Due to availability of partial nad1 and cox1 genes from different geographical locations, phylogenetic analysis was conducted using 677 and 735 bp of the nad1 and cox1 genes, respectively. The Bayesian phylogeny further confirmed identity of the isolates, as they clustered with other reference T. hydatigena sequences from the GenBank database (Supplementary Table S1) and exhibited significant distances from other Taenia species (T. saginata, T. ovis, T. pisiformis, T. solium, T. multiceps, and T. asiatica) (Fig. 2A, B), indicating that all isolates were clearly T. hydatigena. This result was also confirmed by the posterior probability (PP) values between the isolates, although a few had PP values <0.70. This low nodal support value between T. hydatigena species was also recently reported in Nigeria [18]. The nucleotide sequences of cox1 and nad1 genes representing T. hydatigena isolates from the present study were deposited in GenBank under the following accession numbers (cox1: MN114527 and MN114528; nad1: MN 114529–MN114531).
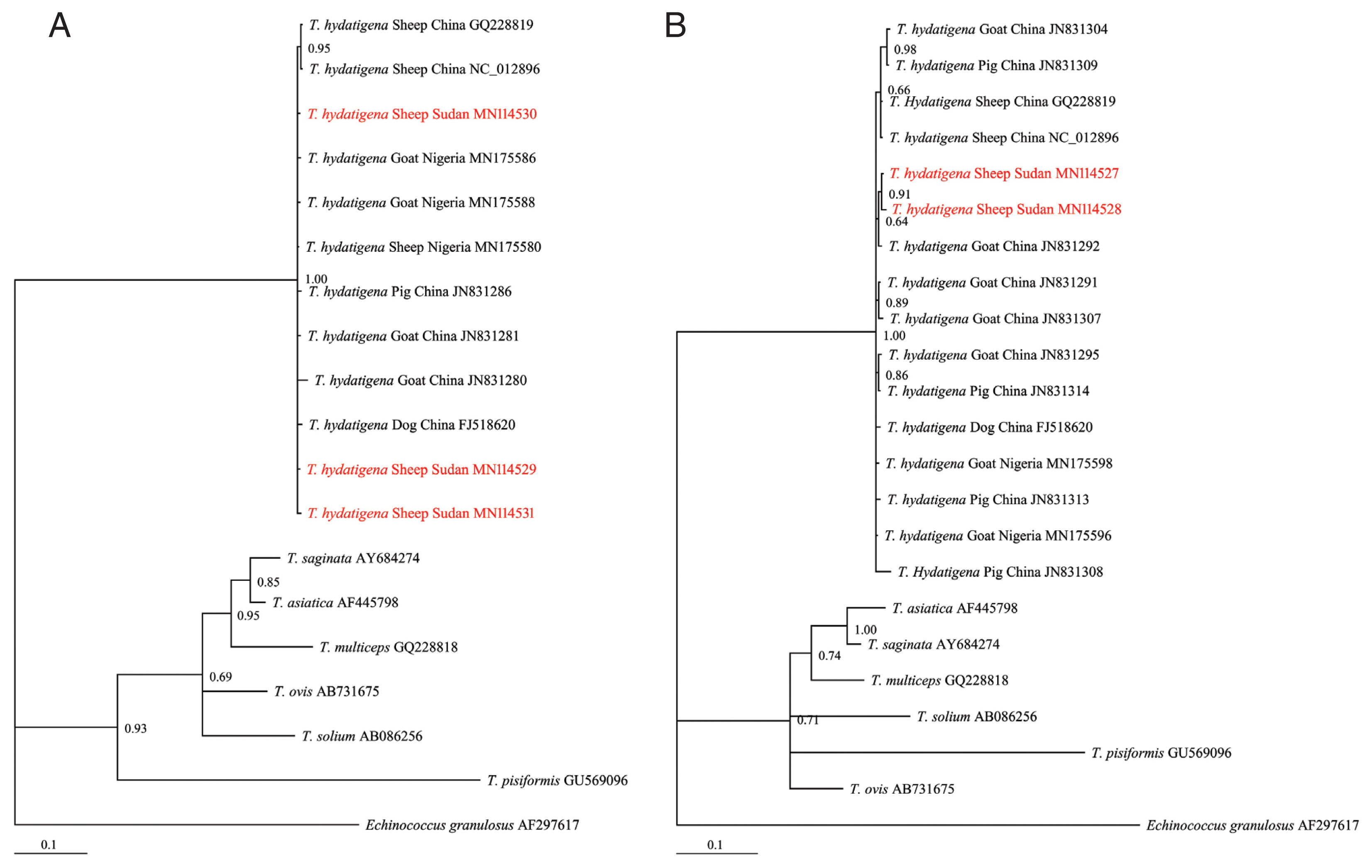
Bayesian phylogenetic relationship of T. hydatigena isolates with other Taenia species. (A) nad1. (B) cox1. Red, Sudanese haplotype representing sheep isolates from this study.
Analysis on the nad1 (764 bp) and cox1 (735 bp) mitochondrial gene sequences revealed 4 (1 parsimony informative) and 2 mutations (not parsimony informative), respectively. No deletions or insertions were observed. Low haplotype (Hd) and nucleotide (π) diversities were observed (Table 1). Regardless of small sample size, the observed nad1 Hd was comparable to T. hydatigena populations reported in dogs and goats, although with a lower π compared to the T. hydatigena populations from the same study [29]. A comparison of the cox1 diversity indices also revealed lower Hd and π than the populations from Italian, Iranian, and Palestinian sheep [29]. Meanwhile, comparable data from an African host on the diversity indices of T. hydatigena are lacking [4,17]. However, compared with the recent report from Nigeria [18], the Sudanese T. hydatigena isolates showed lower genetic variation.
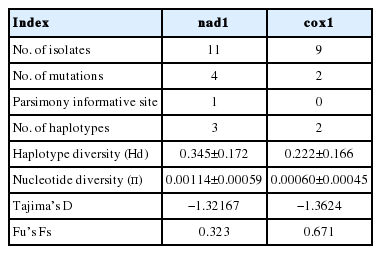
Diversity and neutrality indices of a Taenia hydatigena population from Sudan based on partial nad1 (764 bp) and cox1 genes (735 bp)
Based on the nad1 gene, 3 haplotypes were observed, with the nSDN1 haplotype constituting 81.8% (9/11) of the population. All haplotypes were separated from each other by no more than a 3-point mutational difference. The number of haplotypes based on the cox1 gene was lower compared to the number of haplotypes (4) reported among a T. hydatigena population of Tanzanian sheep and goats [4] but similar to the number of haplotypes reported in Nigerian sheep [18]. The comparison of the nucleotide sequences from this study with the deposited sequences in the GenBank database yielded no haplotype with 100% identity, which could suggest the presence of unique haplotypes in Sudan and is consistent with previous reports of the existence of geographically distinct species of T. hydatigena [29]. Nonetheless, with limited sample size and the use of short mitochondrial DNA (mtDNA) fragments, further investigation would be necessary to understand and clearly describe the genetic differences and relationships between Sudanese T. hydatigena isolates and those from different geographical regions. Furthermore, we observed negative insignificant values of Tajima’s D and insignificant positive Fu’s Fs values (Table 1), indicating inconsistencies with population expansion or a non-significant deviation from neutrality. As expected, although the low sample size may not reflect the actual status of the parasite population in Sudan, it provides a clue to the genetic status of the T. hydatigena population in the country.
The genetic ecology of most parasites of medical and veterinary importance has been found to contribute to understanding the epidemiology and control of parasitic infections. In this study, the results suggest some degree of genetic variation among Sudanese T. hydatigena isolates based on partial nad1 and cox1 mitochondrial genes. While the isolates were highly similar to isolates from other locations, no 100% match was observed. Globally, there still is a dearth of information on the genetic variation of T. hydatigena species, and with few available studies from Africa, this report contributes significant preliminary data on the genetic variation of T. hydatigena species in Sudan. Finally, we recommend that molecular investigations that include large sample sizes and utilize complete mitochondrial gene markers in addition to the nad1 and cox1 genes be considered in the future to provide further insight regarding the genetic variation and population structure of T. hydatigena in Sudan.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Plan (2017YFD0501301, 2017YFD0501302), the Central Public-interest Scientific Institution Basal Research Fund (1610312016012, 1610312017001) and innovation fund 1 of the Chinese Academy of Agricultural Sciences (CAAS), while a scholarship was provided by the Chinese Government Scholarship (CGS) Programme of the Peoples Republic of China for Rosline James Muku.
Notes
The authors declare no conflict of interest related to this study.