Abstract
Interactions between GRA proteins of dense granules in Toxoplasma gondii and host cell proteins were analyzed by yeast two-hybrid technique. The cMyc-GRA fusion proteins expressed from pGBKT7 plasmid in Y187 yeast were bound to host cell proteins from pGADT7-Rec-HeLa cDNA library transformed to AH109 yeast by mating method. By the selection procedures, a total of 939 colonies of the SD/-AHLT culture, 348 colonies of the X-α-gal positive and PCR, 157 colonies of the X-β-gal assay were chosen for sequencing the cDNA and finally 90 colonies containing ORF were selected to analyze the interactions. GRA proteins interacted with a variety of host cell proteins such as enzymes, structural and functional proteins of organellar proteins of broad spectrum. Several specific bindings of each GRA protein to host proteins were discussed presumptively the role of GRA proteins after secreting into the parasitophorous vacuoles (PV) and the PV membrane in the parasitism of this parasite.
-
Key words: Toxoplasma gondii, GRA proteins, parasitophorous vacuole, parasitophorous vacuole membrane, secretion, yeast two-hybrid technique, GRA-host protein interaction, parasitism
INTRODUCTION
Toxoplasma gondii, the causative agent of toxoplasmosis, is an obligate intracellular parasite, which can penetrate actively into any nucleated cells from a wide range of vertebrates including humans. During the entry of
T. gondii, a parasitophorous vacuole (PV) is formed within the host cells and compartmentalized from the host cell cytoplasm by a PV membrane (PVM) within which the parasites multiply and grow. In the toxoplasmal PV and PVM, a variety of proteins are secreted from dense granules and rhoptries, which interact with the cytosolic components and subcellular organelles of the host cells, as evidenced morphologically by the recruitment of the host endoplasmic reticulum and mitochondria (
Magno et al., 2005).
In the dense granule, 10 GRA proteins (GRA1 - GRA10) without sequence homology to each other (
Adjogble et al., 2004;
Ahn et al., 2005;
reviewed by Mercier et al., 2005), 2 isoforms of nucleotide triphosphate hydrolase (NTPase I and II) (
Johnson et al., 2003) and 2 protease inhibitors (TgPI 1 and 2) (
Morris et al., 2002;
Pszenny et al., 2002) have been previously identified in
T. gondii tachyzoites. When secreted into the PV, only GRA1, TgPIs, and the NTPases remain primarily in the lumen of the vacuole (
Sibley et al., 1995;
Pszenny et al., 2002). Most GRA proteins have been detected in close association with the membraneous system of this compartment. Hence, a fraction of the GRA1, GRA2, GRA4, GRA6, GRA7, NTPases pools, GRA8 and GRA9 has been detected more specifically associated with the vacuolar network membranes (
Charif et al., 1990;
Dubremetz et al., 1993;
Sibley et al., 1994;
Lecordier et al., 1995;
Bonhomme et al., 1998;
Labruyere et al., 1999;
Carey et al., 2000;
Adjogble et al., 2004). In these membranes, GRA2, GRA4 and GRA6 participate in the formation of a multimeric protein complex (
Labruyere et al., 1999). In contrast, GRA3, GRA5 and GRA10 are preferentially detected as PVM-associated proteins (
Achbarou et al., 1991;
Lecordier et al., 1993;
Ahn et al., 2005).
Dense granular proteins of known functions such as NTPases and TgPIs have been identified enzymatic properties of the proteins before being confirmed the localization in the dense granules of T. gondii. However, until now the functions of GRA proteins have been restricted to genetic information regarding cDNAs and the domains and/or motifs of the deduced amino acid sequences. Therefore, interactions of secreted GRA proteins in the PV and PVM with host cell proteins and organelles will provide a way to elucidate the functions of GRA proteins in the parasitism of T. gondii. In this study, we profile the host cell proteins that bind to GRA proteins via yeast two-hybrid technique.
MATERIALS AND METHODS
Yeast strains
All vectors, yeast strains, reagents, and methods were adapted from the BD MATCHMAKER™ Library construction & Screening Kit (Clontech, Palo Alto, California, USA). The bacterial strain Escherichia coli DH5α was used for the propagation of the plasmid constructs. The yeast strains Saccharomyces cerevisiae AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) and Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met-, gal80Δ, URA3::GAL1UAS-GAL1TATA-lacZ, MEL1) were employed as hosts in the two-hybrid assay. AH109 contains 2 nutritional reporter genes for adenine and histidine. Both AH109 and Y187 harbor the LacZ reporter gene.
Bait plasmid construction of GRA cDNAs
The RH strain of
T. gondii was maintained by peritoneal passage in Balb/c mice. Whole
T. gondii tachyzoite RNA was extracted using TRIzol Reagent according to the manufacturer's instructions (Gibco BRL, Grand Island, New York, USA). One µg of total RNA was reverse transcribed into cDNA with MMLV reverse transcriptase. Amplification of DNA fragment encoding
T. gondii GRA proteins was achieved by PCR on the cDNA with the primer sets in
Table 1. Each of the DNA fragments was inserted into pGBKT7 after EcoRI/BamHI digestion to transform into Y187 yeast.
In order to construct the cDNA library, total RNA was isolated from HeLa cells using TRIzol Reagent according to the manufacturer's protocol (Gibco BRL). Two µg of total RNA was reverse transcribed into cDNA using CDS III/6 random primers and MMLV reverse transcriptase. After 10 min, the SMART III oligonucleotide was added to the reaction, which resulted in the creation of cDNA flanked by the SMART and CDS sites. The cDNA was amplified by long-distance PCR using the Advantage 2 Polymerase Mix (Clontech), and size-fractionated over a CHROMA SPIN+TE-400 column (Clontech) to reduce fragments smaller than 200 bp. The remaining cDNA was then transformed into competent AH109 yeast along with the target (pGADT7-Rec) vectors, the latter of which is designed to undergo homologous recombination with the corresponding SMART and CDS sites on the ends of the target inserts in vivo to generate fusions to the GAL4 activation domain (AD). The AH109 strain harbors multiple reporter genes (ADE2, HIS3, MEL1 and lacZ) that should only be expressed if the GAL4 DB is brought into proximity to the AD by virtue of an interaction between their bait and the target fusion partners (
Nalani and Sullivan, 2005).
The specificity of interaction was evaluated by yeast mating according to the manufacturer's instructions (Clontech). Briefly, pGBKT7-GRAxx, pGBKT7-Lam, pGBKT7-53 or pGBKT7 vector alone was transfected into Y187 (MATα) and grown in medium lacking tryptophan; pGADT7-Rec-HeLa library or pGADT7-RedT or pGADT7-Rec vector alone was transformed into AH109 (MATa) and grown in medium lacking leucine. The transfected AH109 was then mated to Y187 cells and grown in medium lacking adenine, histidine, tryptophan and leucine. After growing for 10 days, the cells were replica plated in medium lacking adenine, histidine, tryptophan and leucine with X-α-Gal. Y187 transfected with pGBKT7 vector alone or with pGBKT7-Lam was used as a negative control for fortuitous interaction with transfected AH109 with pGADT7-Rec vector alone or with pGADT7-RecT, whereas Y187 transfected with pGBKT7-53 vector was employed as a positive control for definite interaction with transfected AH109 with pGADT7-RecT (
Wong et al., 2002).
The β-galactosidase activity of positive clones subsequently was quantified using a chlorophenol red-β-D-galactopyranoside (CPRG) spectrophotometric assay. In brief, yeast was grown on SD-Ade/Trp/Leu/His plates for 3-5 days at 30℃. Several colonies from each plate were inoculated into 5 ml of liquid SD-Ade/Trp/Leu/His medium and grown overnight at 30℃ with shaking at 250 rpm. From the liquid culture, 2 ml was removed, diluted into 8 ml of YPD medium and grown at 30℃ to mid-log phase (A600 of 0.5-0.8). Optical density at 600 nm was used to normalize β-galactosidase activity to yeast cell density. A 1.5 ml aliquot of each yeast culture was centrifuged briefly to pellet the cells and resuspended in 1 ml of Buffer 1 (2.38 g HEPES, 0.9 g NaCl, 65 mg L-aspartate, 1 g BSA, 50 µl Tween-20 in 100 ml, pH 7.30). Each resuspended yeast culture was centrifuged, and the pellet was resuspended in 0.3 ml of Buffer 1. The resuspension was repeated the freeze/thaw cycles, and β-galactosidase assays were initiated by addition of 0.7 ml of Buffer 2 (27.1 mg CPRG in 20 ml Buffer 1). The reactions were terminated by addition of 0.5 ml of 3 mM ZnCl2, and the reaction mixtures were centrifuged for 1 min at 14,000 rpm. Absorption at 578 nm (A578) was measured for each of the samples and converted into β-galactosidase units. One unit of β-galactosidase is defined as the amount which hydrolyzes 1 µmol of CPRG to chlorophenol red and D-galactose per min per cell. Positive clones with β-galactosidase activities of at least twice the background levels were selected for further characterization.
DNA sequencing and analysis
From the positive colony, the library inserts containing GAL4 AD were amplified by PCR using the upper primer (MATCHMAKER 5'AD LD-Insert Screening Amplimer) 5'-CTA TTC GAT GAT GAA GAT ACC CCA CCA AAC CC-3' and lower primer (MATCHMAKER 3'AD LD-Insert Screening Amplimer) 5'-GTG AAC TTG CGG GGT TTT TCA GTA TCT ACG ATT-3' (Clontech). The amplified DNA fragments were cloned into a vector, pGEM-T Easy vector (Promerga, Madison, Wisconsin, USA). Clones with inserts were selected for sequence analysis using SP6 primer (5'- ATT TAG GTG ACA CTA TAG-3'). Nucleotide sequencing was performed by Macrogen (Seoul, Korea). The presence of open reading frames fused to the GAL4 AD squences was verified and the sequences were searched for homologous sequences with BLAST search analysis (
www.ncbi.nlm.nih.gov/BLAST/) (
Hennig et al., 2001).
RESULTS
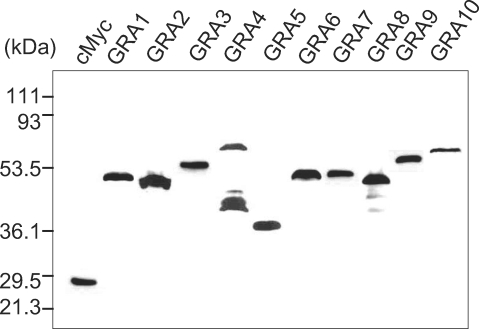
The cMyc fusion proteins of GRAs cloned in the pGBKT7 plasmid were expressed in yeast (Y187) and were blotted with anti-cMyc antibody to confirm the fulfillment of the GRA baits in the yeast two-hybrid system as prey for the HeLa proteins as shown in
Fig. 1. All GRA proteins were expressed well as cMyc fusion proteins, with the exception of the GRA4 of cMyc-GRA4 fusion protein which were detected with minor smudge. After mating the pGBKT7-GRA containing Y187 (MATα) with a pGADT7-Rec-HeLa cDNA library containing AH109 (MATa) positive clones were chosen according to the selection procedures, which resulted in a total of 939 colonies of the SD/-AHLT culture, 348 colonies of the X-α-gal positive and PCR, 157 colonies of the X-β-gal assay and 90 colonies of the ORF search as counted in
Table 2.
Yeast two-hybrid analysis with GRA1 as bait to the prey of the HeLa cDNA library resulted in 8 clones of 7 Homo sapiens genes as 2 Mof4 family associated protein 1 (MRFAP1, NM_033296), Coenzyme A synthase (COASY, NM_025233), laminin beta 3 (LAMB3), transcript variant 1 (NM_000228), ribosomal protein L10a (RPL10A, NM_007104), NAD(P)H dehydrogenase, quinine 2 (NQO2, NM_000904), cofactor required for Sp1 transcriptional activation, subunit 2, 150 kDa (CRSP2, NM_004229) and leukotriene B4 receptor 2 (LTB4R2, NM_019839).
The use of GRA2 as bait for the prey of HeLa cDNA library resulted in no interaction with the host proteins. The use of GRA3 as bait for the prey of the HeLa cDNA library resulted in 8 clones of 4 Homo sapiens genes of 3 calcium modulating ligand (CAMLG, NM_001745), 3 paired box gene 6 (aniridia, keratitis) (PAX6), transcript variant 2 (NM_001604), ribosomal protein S18 (RPS18, NM_022551) and FUN14 domain containing 1 (FUNDC1, NM_173794).
The use of GRA4 as bait for the prey of the HeLa cDNA library resulted in 21 clones of 13 Homo sapiens genes of 6 cofilin 1 (non-muscle) (CFL1, NM_005507), 2 pyruvate kinase (muscle) (PKM2) transcript variant 1 (NM_002654), 2 tRNA splicing endonuclease 34 homolog (S. cerevisiae) (TSEN34, NM_024075), 2 tranlocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) (TIMM50, NM_001001563), tumor susceptibility gene 101 (TSG101, NM_006292), nicotinate phosphoribosyltransferase domain containing 1 (NAPRT1, NM_145201), presenilin enhancer 2 homolog (C. elegans) (PSENEN, NM_172341), WD repeat domain 68 (WDR68) transcript variant 2 (NM_001003725), RNA binding motif protein 9 (RBM9) transcript variant 1 (NM_001031695), thymidine kinase 1, soluble (TK1, NM_003258), MHC class I antigen (HLA-A*0201 allele, AY365426), cortactin (BC033889) and translocase of inner mitochondrial membrane 13 homolog (yeast) (TIMM13, NM_012458). The use of GRA5 as bait for the prey of the HeLa cDNA library resulted in 4 clones of 2 Homo sapiens genes of 3 calcium modulating ligand (CALMG, NM_001745) and small glutamine-rich tetratricopeptide repeat (TRP)-containing alpha (SGTA, NM_003021).
The use of GRA6 as bait for the prey of the HeLa cDNA library resulted in 8 clones of 4 Homo sapiens genes of 5 calcium modulating ligand (CAMLG, NM_001745), spectrin repeat containing nuclear envelope 2, transcript variant 1 (SYNE2, NM_015180), ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle (ATP5A1), nuclear gene encoding mitochondrial protein, transcript variant 2 (NM_004046), and proteasome (prosome, macropain) subunit, alpha type 4 (PSMA4, NM_002789). The use of GRA7 as bait for the prey of the HeLa cDNA library resulted in 2 Homo sapiens genes of poly (rC) binding protein 1 (PCBP1, NM_006196) and thymosin beta 10 (TMSB10, NM_021103).
The use of GRA8 as bait for the prey of the HeLa cDNA library resulted in 24 clones of 14 Homo sapiens genes of 4 thymidine kinase 1, soluble (TK1, NM_003258), 4 RNA binding motif protein 9 (RBM9) transcript variant 1 (NM_001031695), 2 nucleotide binding protein 2 (MinD homolog, E. coli) (NUBP2, NM_012225), hydroxyacyl-Coenzyme A dehydrogenase type II (HADH2), nuclear gene encoding mitochondrial protein, transcript variant 1 (NM_004493), actinin alpha 1 (ACTN1, NM_001012), ATP synthase, H+ transporting mitochondrial F0 complex subunit C1 (subunit 9) (ATP5G1), nuclear gene encoding mitochondrial protein, transcript variant 1 (NM_005175), ribosomal protein SA, transcript variant 1 (RPSA, NM_002295), phosphoglyucerate dehydrogenase (PHGDH, NM_006623), ribosomal protein L10 (RPL10, NM_006013), nitrilase family member 2 (NIT2, NM_020202), cadherin-like 24, transcript variant 1 (CDH24, NM_022478), eukaryotic translation initiation factor 3 subunit 2 beta, 36 kDa (EIF3S2, NM_003757), pyruvate kinase, muscle, transcript variant 3 (PKM2, NM_182471) and cytochrome b5 reductase 3, transcript variant M (CYB5R3, NM_000398). The use of GRA9 as bait for the prey of the HeLa cDNA library resulted in 4 clones of 3 Homo sapiens genes of 2 filamin B beta (actin binding protein 278) (FLNB, NM_001457), metallothionein 2A (MT2A, NM_005953) and processing of precursor 7 ribonuclease P subunit (S. cerevisiae) (POP7, NM_005837).
The use of GRA10 as bait for the prey of the HeLa cDNA library resulted in 11 clones of 7 Homo sapiens genes of 3 NADH dehydrogenase subunit 1 (AAW58628), 3 HSPC009 protein (BC002698), HLA-B associated transcript 8 (BAT8) isoform a variant protein (AB209433), signal transducer and activator of transcription 6, interleukin-4 induced (STAT6, NM_003153), TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63 kDa (TAF1B, NM_005680), solute carrier family 10 (sodium/bile acid cotransporter family) member 3 (SLC10A3, NM_019848) and RNA binding protein 1 (RANBP1, NM_002882).
DISCUSSION
GRA proteins interacted with a number of host cell proteins including enzymes, and structural and functional subcellular organellar proteins of broad spectrum by yeast two-hybrid methods, with some specific pattern to each GRA protein. PV formation and the structural modifications of PVM between the parasite and the host cells have been studied extensively in apicomplexan parasitism, especially in
Plasmodium falciparum and
T. gondii. Among the 3 secretory organelles of the apicomplexan parasites, rhoptry proteins are major secreted proteins to the plasmodial PVM, which interact with the host components and/or sometimes are secreted to the cytosol across the PVM, and reach to the surface membrane via the tubulovesicular membrane (TVM) network of the relatively simple host cells with few subcellular organelles, erythrocytes (
Templeton and Deitsch, 2005). On the while,
T. gondii infects all nucleated host cells including macrophages, which encounter a great deal of complex and various interactions with host cell components and subcellular organelles in toxoplasmal PVM. Secreted dense granular proteins are thought to be in charge of these interactions, in addition to rhoptry proteins. The exocytosed dense granular proteins either remain soluble in the lumen of the PV or become associated with the PVM or the tubulovesicular network (TVN) of the membraneous structure within the PV (
Mercier et al., 2002), which are thought to modify the environment within the PV for interaction with host components, allowing for functions in intracellular survival and replication (
Lingelbach and Joiner, 1998).
GRA3, GRA5 and GRA6 are commonly bound to the calcium modulating ligand (CAMLG). CAMLG is an integral membrane protein which appears to be a new participant in the calcium signal transduction pathway. CAMLG functions in a similar fashion on to cyclosporin A, binding to cyclophilin B and operating downstream of the T-cell receptor (TCR) (
Guo et al., 2005) and upstream of calcineurin (
Bram and Crabtree, 1994) via the induction of a calcium influx. The modulation of intracellular calcium concentrations by these GRA proteins bound to CAMLG leads to the inhibition of host cell apoptosis (
Feng et al., 2002) for the long-term residence of the intracellular parasites. Structurally, GRA3, GRA5 and GRA6 bound to CAMLG in the intracellular integral membrane of ER, which has been suggested as the ligand of ER anchorage to PVM during the parasitism of
T. gondii. GRA3 and GRA5 are secreted into the PVM to coordinate structurally in binding to CAMLG in the intracellular integral membrane of ER, and are linked to GRA6 inside the PV.
GRA4 binds to cofilin, a widely distributed intracellular actin-modulating protein which binds and deploymerizes filamentous F-actin and also inhibits the polymerization of monomeric G-actin in a pHdependent manner (
Pope et al., 2004;
Vardouli et al., 2005). It is involved in the translocation of the actincofilin complex from the cytoplasm to the nucleus (
Nebl et al., 1996). The GRA4-cofilin interaction may exert to the maintenance of the intravacuolar network. After the induction of apoptosis, cofilin is translocated to the mitochondria prior to the release of cytochrome c. The reduction of cofilin with siRNA results in the inhibition both of cytochrome c release and apoptosis (
Chua et al., 2003). Of course, the localization of GRA4 within the PV, as well as its intervening action with cytoplasmic cofilin to TIMM50 (
Guo et al., 2004) and TIMM13 (
Paschen et al., 2000;
Jensen and Dunn, 2002) from approaching mitochodria to the PVM is associated with the anti-apoptotic activities of these complexes.
GRA5 also binds to SGTA, which is expressed ubiquitously as a housekeeping function in interactions with the 70 kDa heat shock protein (
Angeletti et al., 2002) and HSP90β (
Yin et al., 2006). And GRA6 also interacts with ATP5A1 which is a subunit of mitochondrial ATP synthase that catalyzes the synthesis of ATP (
Moser et al., 2001). GRA6 binds to the proteasome subunit, α type, 4 (PSMA4). Proteasomes, multicatalytic proteinase complexes, are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent manner in a non-lysosomal pathway (
Coux et al., 1996;
Apcher et al., 2004). An essential function of a modified proteasome, the immunoproteasome, is involved in the processing of class I MHC peptides.
GRA7, which binds to PCBP1 along with PCBP2 and hnRNPK, corresponds to the principal cellular poly (rC)-binding proteins (
Pickering et al., 2004). It contains 3 K-homologous (KH) domains which might be involved in RNA binding (
Leffers et al., 1995). PCBP1 has also been suggested to play a part in the formation of a sequence-specific α-globin mRNP complex which is associated with the stability of α-globin mRNA (
Kiledjian et al., 1995).
GRA10 evidenced a peculiar binding pattern of those proteins associated with nuclear and nucleolar localizations. Among these, STAT6 is a member of the STAT family of transcription factors. In response to cytokines and growth factors, the members of the STAT family are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers which translocate into the cell nucleus, in which they function as transcription activators. STAT6 plays a central role in the exertion of IL-4 mediated biological responses. It has been shown to induce the expression of Bcl2L1/Bcl-x(L), which is responsible for the antiapoptotic activity of IL-4 (
Wurster et al., 2002). It functions in the differentiation of T helper 2 (Th2) cells, the expression of cell surface markers and the switch of immunoglobulin classes (
Blanchard et al., 2005). GRA10 also binds to TAF1B. The initiation of transcription by RNA polymerase I requires the formation of a complex composed of the TATA-binding protein (TBP) and 3 TBP-associated factors (TAFs) specific to RNA polymerase I. This complex, known as SL1, binds to the core promoter of the ribosomal RNA genes to position the polymerase properly, and also functions as a channel for regulatory signals (
Comai et al., 1994). And Ran/TC4-binding protein, RanBP1, interacts specifically with GTP-charged Ran. RanBP1 binds to Ran complexed with GTP but not GDP (
Coutavas et al., 1993). RanBP1 does not activate the GTPase activity of Ran, but does markedly increase GTP hydrolysis by the RanGTPase-activating protein (RanGAP1) (
Seewald et al., 2003). RanBP1 functions as a negative regulator of RCC1 by the inhibition of RCC1-stimulated guanine nucleotide release from Ran (
Steggerda and Paschal, 2000). For the binding of GRA10 with host cell nuclear proteins GRA10 must be secreted across the PVM. But, until now, no
T. gondii proteins have been shown to be secreted over the PVM into the host cell cytoplasm or surface membrane, differently from what has been observed in the case of
Plasmodium infection of erythrocytes. Toxoplasmal PVM itself is not a rigid mold, but is rather a biomembrane which provides for endocytosis/exocytosis between the parasite and the host cell cytoplasm. With 2 transmembrane domains and a nuclear localization sequence (NLS), GRA10 deserves a candidate protein that can follow the host cell membrane trafficking to the nucleus as described in
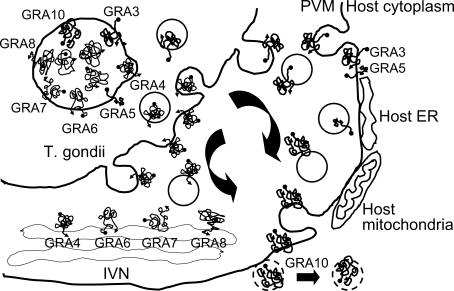
Fig. 2.
Proteins that bind to GRA1, GRA8 and GRA9 are not discussed herein due to the diversity inherent to these binding proteins. Several listed prey proteins were immunoprecipitated with their bait GRA proteins in the preliminary assay (data not shown). Although more specific binding should be confirmed further to analyze the functions of each of the GRA proteins, the results of this trial delineate the preliminary interactions of secreted GRA proteins and host cell proteins for the intracellular parasitism of T. gondii.
Notes
-
This study was supported by the Academic Research Service Program of the Korean National Institute of Health, 2006.
References
Fig. 1The cMyc fusion proteins of GRAs cloned in pGBKT7 plasmid expressed in yeast (Y187). They were blotted by anti-cMyc antibody to verify the fulfillment of the GRA proteins as bait in the yeast two-hybrid system.

Fig. 2Secretion of GRA proteins with transmembrane domains within the amino acid sequences. GRA4, 6, 7 and 8 are trapped in the IVN within the PV, and participate in the interaction with host components across the PVM, whereas GRA3, 5 and 10 are secreted into the PVM for the direct interaction. GRA10 may be a candidate that is secreted over the PVM and interacts with host nuclear proteins.

Table 1.Primer sets used for the amplification of GRA proteins
Table 1.
|
Bait |
Primer |
bp |
|
GRA1 |
5´-CAGAATTCATGGTGCGTGTGAGCGCT-3´ |
573 |
|
5´-ACGGATCCTTACTCTCTCTCTCCTGTTAGGAAC-3´ |
|
|
GRA2 |
5´-GCGAATTCATGTTCGCCGTAAAACATTGT-3´ |
814 |
|
5´-AGGGATCCTTACTGCGAAAAGTCTGGGAC-3´ |
|
|
GRA3 |
5´-GAGAATTCATGGACCGTACCATAT-3´ |
669 |
|
5´-GGGATCCTCAGGTTTGTTTCTTG-3´ |
|
|
GRA4 |
5´-TTGAATTCATGCAGGGCACTTGGTTTTC-3´ |
1038 |
|
5´-CAGGATCCTCACTCTTTGCGCATTCTTTC-3´ |
|
|
GRA5 |
5´-GTGAATTCATGGCGTCTGTAAAACGCG-3´ |
363 |
|
5´-AGGGATCCTTACTCTTCCTCGGCAACTTC-3´ |
|
|
GRA6 |
5´-TCGAATTCATGGCACACGGTGGC-3´ |
693 |
|
5´-GCGGATCCCCCCTGTTTTCATCTTTAATA-3´ |
|
|
GRA7 |
5´-TGAATCCATGGCGGCACACGCAATT-3´ |
746 |
|
5´-CCGGATCCCCTACTGGCGGGCAT-3´ |
|
|
GRA8 |
5´-GTGAATTCATGGCTTTACCATTGCGTGTT-3´ |
804 |
|
5´-CCGGATCCTTAATTCTGCGTCGTTACG-3´ |
|
|
GRA9 |
5´-GGAATCCATGCGGTCACTCAAGTCAATCG-3´ |
974 |
|
5´-CGGGTCGACCTCAGAGTCCTCGGTCTT-3´ |
|
|
GRA10 |
5´-GGCCATATGATGATTGAGGCCGCTGTG-3´ |
1095 |
|
5´-CTGGAATTCTCAGACAGGCGTTTCCCC-3´ |
|
Table 2.Number of positive colonies by the selection procedures
Table 2.
|
Biat |
SD/-AHLT |
X-α-gal & PCR |
X-β-gal |
ORF search |
|
GRA1 |
10 |
9 |
8 |
8 |
|
GRA2 |
5 |
0 |
0 |
0 |
|
GRA3 |
281 |
71 |
8 |
8 |
|
GRA4 |
86 |
57 |
37 |
21 |
|
GRA5 |
17 |
6 |
6 |
4 |
|
GRA6 |
21 |
13 |
10 |
8 |
|
GRA7 |
6 |
3 |
2 |
2 |
|
GRA8 |
433 |
140 |
49 |
24 |
|
GRA9 |
27 |
6 |
4 |
4 |
|
GRA10 |
53 |
43 |
33 |
11 |