Comparative Microbiome Analysis of Three Species of Laboratory-Reared Periplaneta Cockroaches
Article information
Abstract
Cockroaches inhabit various habitats, which will influence their microbiome. Although the microbiome can be influenced by the diet and environmental factors, it can also differ between species. Therefore, we conducted 16S rDNA-targeted high-throughput sequencing to evaluate the overall bacterial composition of the microbiomes of 3 cockroach species, Periplaneta americana, P. japonica, and P. fuliginosa, raised in laboratory for several generations under the same conditions. The experiments were conducted using male adult cockroaches. The number of operational taxonomic units (OTUs) was not significantly different among the 3 species. With regard to the Shannon and Pielou indexes, higher microbiome values were noted in P. americana than in P. japonica and P. fuliginosa. Microbiome composition was also evaluated, with endosymbionts accounting for over half of all OTUs in P. japonica and P. fuliginosa. Beta diversity analysis further showed that P. japonica and P. fuliginosa had similar microbiome composition, which differed from that of P. americana. However, we also identified that P. japonica and P. fuliginosa host distinct OTUs. Thus, although microbiome compositions may vary based on multiple conditions, it is possible to identify distinct microbiome compositions among different Periplaneta cockroach species, even when the individuals are reared under the same conditions.
INTRODUCTION
Insect microbiomes affect the nutrient cycling, provide protection from parasites and pathogens, and modulate immune responses. Cockroach microbiomes consist of horizontally transmitted microbes and vertically transmitted symbionts. The diversity of these microbiomes can vary depending on developmental stage, diet, and rearing practices [1]. In a recent study, laboratory-reared and field-collected Blattella germanica presented distinct microbiomes, although they shared the same core bacterial taxa, which appear to differ depending on the location and diet [2]. However, no significant microbiome differences resulting from changes in diet have been observed in Periplaneta americana, although this species has been found to present microbiome differences due to environmental factors [3].
Forty-seven species are included in the genus Periplaneta [4], and we maintain 3 species of the genus Periplaneta in our laboratory, i.e., P. americana, P. japonica, and P. fuliginosa. P. americana originated in Africa and is very common worldwide [4]. This species measures approximately 4 cm in length [7] and is often found in commercial buildings [5]. Periplaneta fuliginosa is another species of African origin and measures 3 cm in length. This species is widely distributed across the southeastern United States and Japan [6]. Periplaneta japonica, which measures 2.5 cm in length, originated in the Japan and is freeze tolerant [7].
As the features of each species differ within the genus Periplaneta, we expected that there would also be differences among Periplaneta microbiomes. Hence, we conducted a study to establish a microbiome that minimized the aforementioned differences, which may have been because of diet and environmental factors. The laboratory-reared cockroaches used in this study were reared for many generations under the same laboratory conditions to minimize the differences resulting from diet and environmental factors. Then, we analyzed the cockroach microbiomes using 16S rDNA-targeted high-throughput sequencing to compare the 3 cockroach species.
MATERIALS AND METHODS
Cockroach collection and rearing conditions
Individuals of 3 species of cockroaches, P. americana, P. japonica, and P. fuliginosa, were collected in Yongsan, Seoul, and Incheon, respectively. P. americana and P. fuliginosa individuals have been maintained in the laboratory of the Arthropods of Medical Importance Bank of Yonsei University College of Medicine in Seoul, Korea, since 1998, while P. japonica individuals have been reared since 2017. The cockroaches used in this study were reared for many generations under the same laboratory conditions to minimize the potential influence of environmental factors and diet. In addition, all the cockroaches used in this study were in the adult stage. A total of 40 cockroaches were reared separately in a plastic box (27×34×19 cm) maintained at 25˚C. The cockroaches were fed Purina Rat Chow (Nestle Purina PetCare, St Louis, Missouri, USA), containing crude protein, crude fat, crude fiber, crude ash, calcium, and phosphorus, and were supplied tap water ad libitum. Male adult cockroaches (not non-reproductive), one month after the last instar, were used. Sampling was performed 2 days after starving.
DNA extraction
The surface of each cockroach was sterilized using alcohol. The cockroaches (n=6 of each species) were then frozen with liquid nitrogen and individually crushed using a mortar and pestle, and their DNA was extracted. Total DNA was extracted using the NucleoSpin DNA Insect Kit (Macherey-Nagel, Düren, Germany) following the instructions of the manufacturer. The DNA extracted from each sample was eluted in 20 μl of elution buffer. All processing and sequencing procedures were conducted at a clean bench, under a sterilized hood, and in a DNA-free room. DNA concentrations were quantified using an ND-1000 Nanodrop system (Thermo-Fisher Scientific, Waltham, Massachusetts, USA). The extracted DNA was stored at −80°C in a deep freezer.
Next-generation sequencing
The 16S rDNA V3–V4 region was amplified by PCR using forward (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and reverse primers (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) [8,9], utilizing an Illumina MiSeq V3 cartridge (San Diego, California, USA) with 600 cycles. A limited-cycle amplification step was performed to add multiplexing indices and Illumina sequencing adapters. The libraries were normalized, pooled, and sequenced on the Illumina MiSeq V3 cartridge platform following the manufacturer’s instructions.
Bioinformatics and statistics
Bioinformatics analyses were performed following the methods described in our previous study [8]. The EzBioCloud database (https://www.ezbiocloud.net/) [8] was used for the taxonomic assignment using BLAST 2.2.22, and pairwise alignments were generated to calculate the similarity [10,11]. All the described analyses were performed using BIOiPLUG, a commercially available ChunLab bioinformatics cloud platform for microbiome research (https://www.bioiplug.com/) [8]. The reads were normalized to 43,000 to perform the analyses. Determination of Shannon, phylogenetic, and Pielou indexes; unweighted pair group method with arithmetic mean (UPGMA) clustering; principal coordinates analysis (PCoA); permutational multivariate analysis of variance (PERMANOVA); linear discriminant analysis (LDA); and effect size (LEfSe) analysis were performed according to the previous study [8].
RESULTS
The average numbers of read counts assigned to P. americana, P. japonica, and P. fuliginosa were 49,905 reads corresponding to 897 operational taxonomic units (OTUs), 56,565 reads corresponding to 955 OTUs, and 58,013 reads corresponding to 878 OTUs, respectively. The rarefaction curve of all the samples formed a plateau (Supplementary Fig. S1). The number of OTUs was not significantly different among the 3 cockroach species. There were no significant differences in the phylogenetic diversity between species, although it was higher for P. americana than for P. japonica as well as P. fuliginosa. In contrast, the Pielou and Shannon index values were significantly higher for P. americana than for P. japonica and P. fuliginosa (Fig. 1, P=0.004).
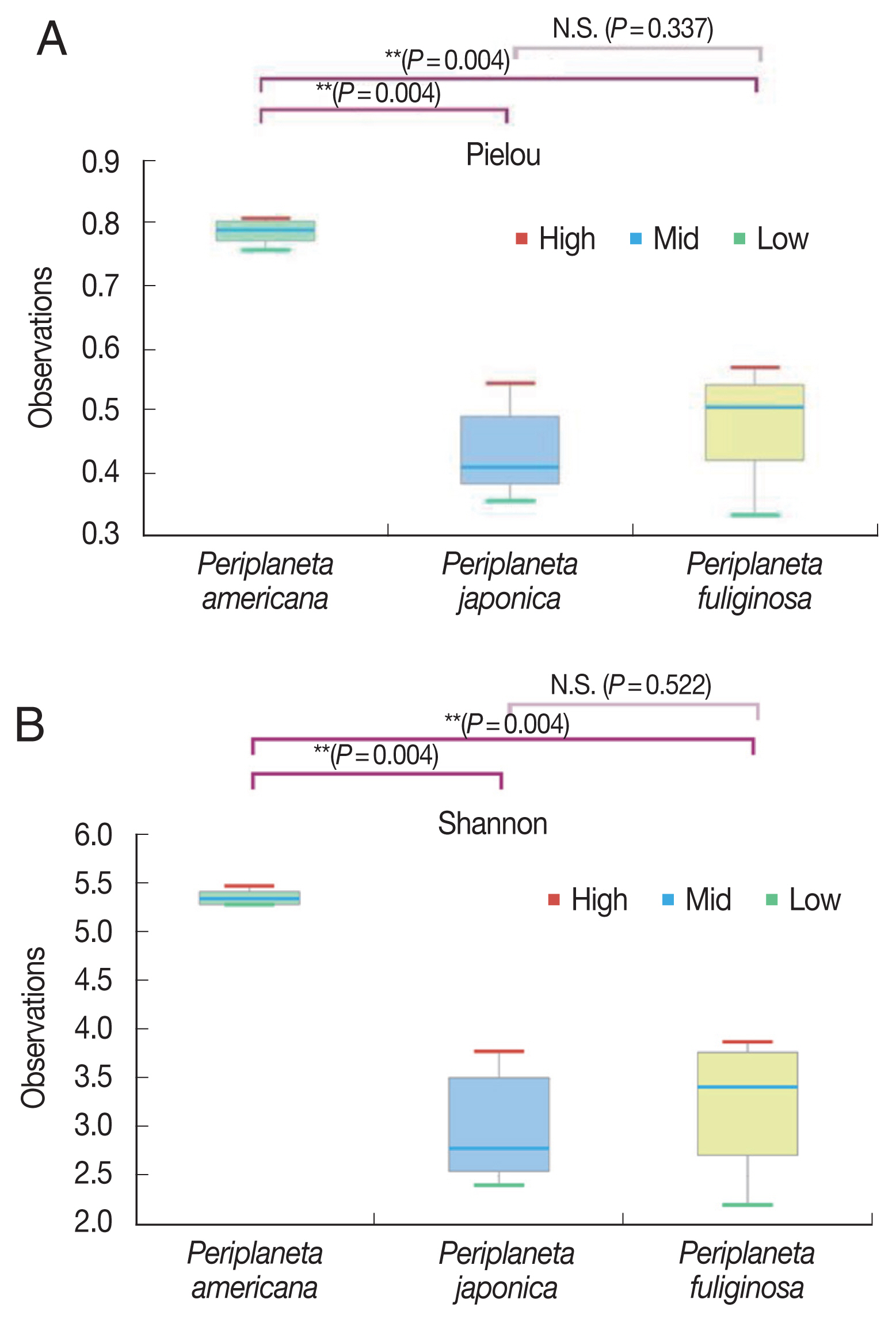
Box plots showing alpha diversity (measurement of species richness, abundance, and equity within a habitat unit) of Periplaneta americana, Periplaneta japonica, and Periplaneta fuliginosa. (A) Pielou diversity (equity). (B) Shannon diversity. Bar indicates median, and hinge represents lower and upper quartiles. *Kruskal-Wallis test, P<0.005.
UPGMA cluster analysis showed that the cockroaches were organized according to species: P. japonica and P. fuliginosa were clustered earlier and then joined the P. americana (Fig. 2A). The results of PCoA showed that although all 3 groups clustered together, P. americana samples were more tightly clustered than the P. japonica or P. fuliginosa samples were (P=0.001) (Fig. 2B). Moreover, a significant difference among the 3 cockroach species with respect to microbiome composition was detected using PERMANOVA [14].

(A) Unweighted pair group method with arithmetic mean clustering. (B) Principal coordinates analysis depicting differences in the taxonomic compositions of the bacterial communities among Periplaneta americana, Periplaneta japonica, and Periplaneta fuliginosa.
With respect to the bacterial taxa present in the 3 cockroach species at the species level (Supplementary Data Set S1), less than 1% of the OTUs in P. americana accounted for 57.1% of all the microbial species present in P. americana. However, bacterial species not included in the aforementioned 1% were more abundant in P. americana than in either P. japonica or P. fuliginosa. The endosymbiont Blattabacterium CP001429_s accounted for 63.1% and 57.3% of all the OTUs in P. japonica and P. fuliginosa, respectively (Fig. 3A). In P. japonica, the endosymbiont Blattabacterium_uc was also present. P. americana lacked Blattabacterium. P. japonica and P. fuliginosa had many OTUs in common, and presented similar microbial species compositions (Fig. 3A).
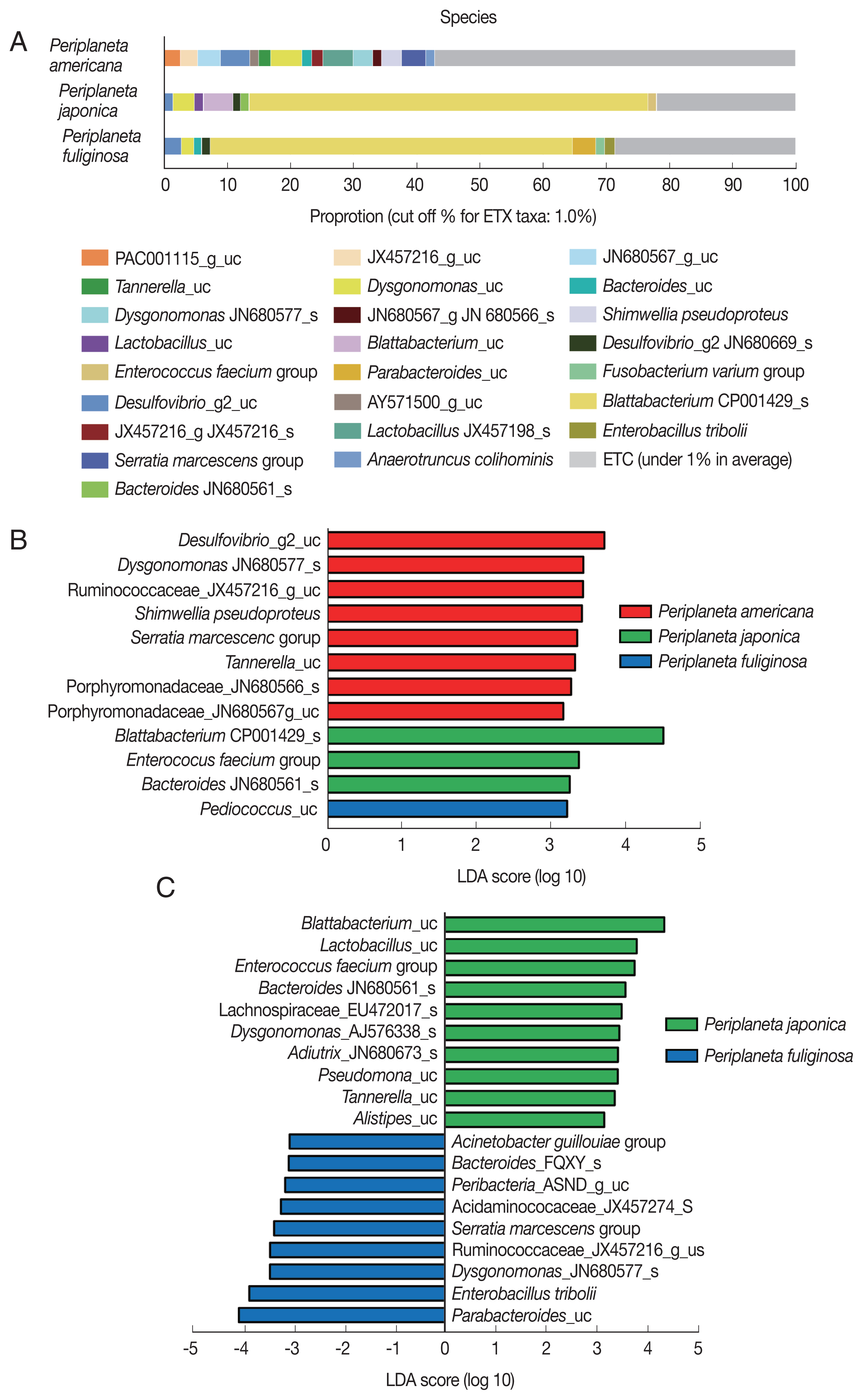
(A) Distribution of bacterial taxa in the 3 cockroach species, Periplaneta americana, P. japonica, and P. fuliginosa. Each bar depicts mean relative abundance value of the independent replicates (n=6). (B) LEfSe analysis of differentially abundant bacterial taxa among the 3 cockroach species, and (C) between P. japonica and P. fuliginosa. Only taxa meeting a significant LDA threshold (>3) are shown.
An LEfSe analysis was performed to identify significant differences in bacterial abundance among the cockroach species. The taxa with high LDA scores in P. americana were Desulfovibrio_g2_uc, Dysgonomonas_JN680577_s, and Serratia marcescens. In P. japonica, Blattabacterium_CP001429_s and Enterococcus faecium presented high LDA scores. Finally, Pediococcus_uc was the species with the highest LDA score in P. fuliginosa (Fig. 3B). When the bacterial communities in P. japonica and P. fuliginosa were compared without P. americana, Blattabacterium_uc and Lactobacillus_uc were found to be highly abundant in P. japonica, while Parabacteroides_uc and Enterobacillus tribolii were highly abundant in P. fuliginosa, and were the species with the highest LDA scores (Fig. 3C).
DISCUSSION
Previous studies have shown that microbiomes may differ based on the diets or rearing conditions of their hosts [1]. The cockroaches used in this study had lived for many generations under the same conditions, and it was, therefore, assumed that other variables, such as diet, temperature, or humidity, would not strongly affect the microbiome.
We evaluated the microbiomes of the 3 cockroach species to determine whether there was any difference among their bacterial profiles. The results indicated that species richness did not differ between cockroach species, but abundance and equity were higher in P. americana than in P. japonica or P. fuliginosa. This means that the richness is the same with the same food and environment, but abundance and equity are expected to be the highest in P. americana, which is the largest in size.
A previous study had shown that the microbiome of P. americana was resilient and stable when the cockroach underwent a dietary shift [3]. In addition, no significant phylum-level differences were reported in the observed microbiomes among the 3 P. americana groups (i.e., diet with 6 foods, mixed diet, and starvation diet). Furthermore, no differences were found among the 3 P. americana groups with respect to either alpha or beta diversity, although differences in microbiome composition that were attributable to environmental factors were observed [3]. Similarly, in this study, assuming P. americana is stable with regard to dietary shifts, we observed the differences in microbial composition attributable to environmental factors at the phylum level (Supplementary Fig. S2).
With respect to the species composition, OTUs (over 1%) in P. americana accounted for the greatest diversity among the 3 cockroach species. Previous studies reported the presence of Blattabacterium in P. americana, contrasting with the findings on our laboratory-reared individuals [13]. However, despite the lack of Blattabacterium, this strain had no problem with reproduction or growth. In a previous study, wood-feeding dictyopteran insects had significantly reduced the numbers of Blattabacterium, resulting in nutrients gained by other means [14]. Therefore, it is possible that the microbiome of the P. americana strain reared in our laboratory may have adapted to the laboratory-related environmental conditions. In P. japonica and P. fuliginosa, Blattabacterium CP001429_s accounted for more than half of all OTUs. In addition, we observed that many OTUs were present in the microbiomes of P. japonica and P. fuliginosa. Nonetheless, the differences among the 3 cockroach species with respect to OTU composition were identified from the results of the UPGMA and PCoA clustering analyses. We confirmed that P. japonica and P. fuliginosa clustered before P. americana. In combination with the species composition results, these results suggest that P. japonica and P. fuliginosa have more similar bacterial compositions compared to that of P. americana. As mentioned earlier, P. americana remained resilient and stable despite a dietary shift [3], and P. japonica and P. fuliginosa are expected to have a similar bacterial composition because of the same diet.
P. americana exhibited higher abundance than P. japonica and P. fuliginosa. We expected that the species were more prevalent because P. americana is larger than P. japonica or P. fuliginosa. Dysgonomonas species, which were present in P. americana, may cause gastroenteritis in immunocompromised humans [15]. Dysgonomonas is common in cockroaches [18], and unclassified Dysgonomonas species were also present in P. japonica and P. fuliginosa. Serratia marcescens has been found to be associated with hospital-acquired infections and is an opportunistic pathogen that is often involved in urinary tract and wound infections [17]. Blattabacterium_CP001429_s was present in P. fuliginosa, but it was more specific to P. japonica. Enterococcus faecium can live in the gastrointestinal tract of both humans and animals, although it can cause endocarditis [18]. P. fuliginosa had many OTUs in common with P. japonica, but Pediococcus_uc was more abundant in all 3 cockroach species. Nevertheless, the PCoA and UPGMA analyses showed different clustering results, and P. fuliginosa showed a substantial number of OTUs in common with P. japonica, although differences were found between the 2.
In future studies, differences between P. japonica and P. fuliginosa with regard to diet and environmental shifts should also be assessed to determine which species characteristics are determined by bacteria. Furthermore, this information will be beneficial for identifying species-specific cockroach features.
In conclusion, we compared the microbiomes of 3 Periplaneta species and found differences in the bacterial composition of their microbiomes despite being reared under the same conditions for many generations.
Supplementary Information
ACKNOWLEDGMENT
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (MEST; numbers NRF-2019R1A2B5B01069843 and 2020R1I1A2074562).
Notes
CONFLICT OF INTERST
The authors have no conflicts of interest to declare.
