Functional Identification of a Nuclear Localization Signal of MYB2 Protein in Giardia lamblia
Article information
Abstract
MYB2 protein was identified as a transcription factor that showed encystation-induced expression in Giardia lamblia. Although nuclear import is essential for the functioning of a transcription factor, an evident nuclear localization signal (NLS) of G. lamblia MYB2 (GlMYB2) has not been defined. Based on putative GlMYB2 NLSs predicted by 2 programs, a series of plasmids expressing hemagglutinin (HA)-tagged GlMYB2 from the promoter of G. lamblia glutamate dehydrogenase were constructed and transfected into Giardia trophozoites. Immunofluorescence assays using anti-HA antibodies indicated that GlMYB2 amino acid sequence #507–#530 was required for the nuclear localization of GlMYB2, and this sequence was named as NLSGlMYB2. We further verified this finding by demonstrating the nuclear location of a protein obtained by the fusion of NLSGlMYB2 and G. lamblia glyceraldehyde 3-phosphate dehydrogenase, a non-nuclear protein. Our data on GlMYB2 will expand our understanding on NLSs functioning in G. lamblia.
Giardia lamblia is a protozoan, which completes its life cycle in 2 forms, namely trophozoite and cyst. Differential display reverse transcription-PCR, in conjunction with in vitro encystation, allowed us to identify G. lamblia MYB2 (GlMYB2, GiardiaDB GL50803_8722) with encystation-induced expression [1]. Independently, it was also identified as a myb-like gene in Giardia genome database searches, and its expression was increased during encystation [2]. The deduced GlMYB2 amino acid sequences indicated that GlMYB2 functioned as a transcription factor with a DNA-binding domain, which comprised 2 imperfect repeats at its carboxyl-terminus (C-terminus). GlMYB2 should be localized into the nucleus of G. lamblia to function as a transcription factor. The nuclear localization of GlMYB2 during encystation was observed in vivo via the expression of a GlMYB2-GFP fusion protein or epitope-tagged GlMYB2 [1,2]. The binding sites of this transcription factor were found in encystation-induced promoters of G. lamblia cyst wall protein 1 (GlCWP1) and GlMYB2 via a random site selection experiment and subsequent gel shift assays [1].
Nuclear localization signals (NLSs) are the specific amino acid sequences of eukaryotic nuclear proteins required for nuclear import of the proteins via nuclear pore complexes [3]. The study on the NLS of G. lamblia included 65 Giardia strains that expressed C-terminal GFP-tagged proteins, which demonstrated nuclear localization of proteins derived from the ventral disc [4]. Analysis of these candidate nuclear proteins was performed using the NLS prediction software NLStradamus [5] with the 2-state HMM static model and posterior prediction with a cutoff of 0.6. Three candidate NLSs were used to demonstrate the expression of the Streptococcus pyogenes Cas9 protein, which includes the 34-amino acid C-terminal NLS from the Giardia protein GL50803_2340, in Giardia nucleus [6].
In this study, we performed analysis of GlMYB2 NLSs. Nuclear localization of GlMYB2 has been confirmed in previous studies [1,2]. The NLS prediction software NLStradamus [5] with the 2-state HMM static model and posterior prediction with a cutoff of 0.1 revealed 2 putative NLSs in GlMYB2 (amino acid sequences #461-#498 and #507-#530). Three NLSs [the 2 NLSs found in the previous analysis using NLStradamus with an additional GlMYB2 NLS (amino acid sequence #334-#359)] were also predicted using cNLS mapper (available at http//nls-mapper.iab.keio.ac.jp/) with a cutoff of 0.3 (Fig. 1A).
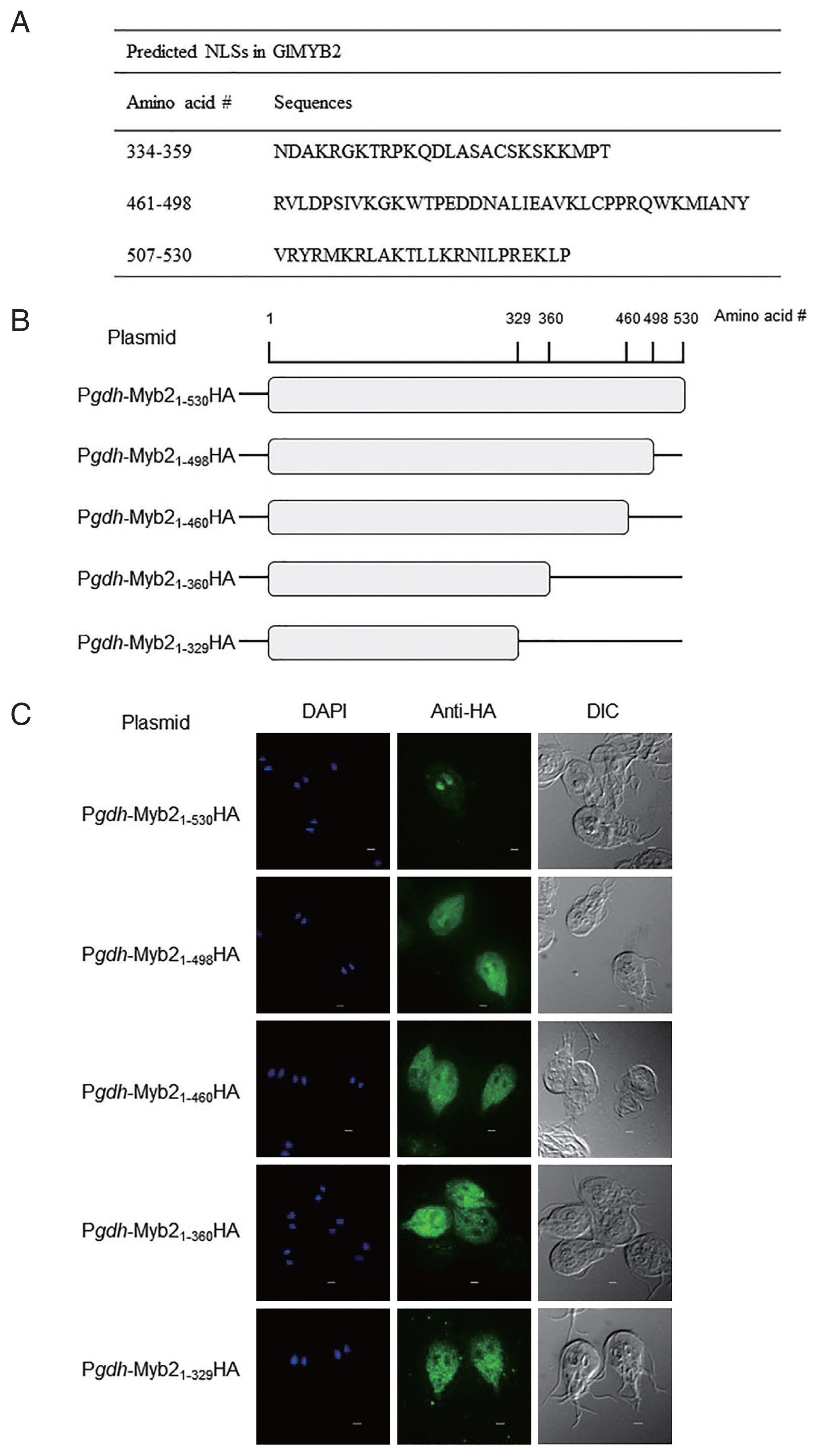
Prediction of nuclear localization signals (NLSs) of G. lamblia MYB2 (GlMYB2) and their role in nuclear localization of GlMYB2 in Giardia. (A) List of putative GlMYB2 NLSs predicted by the NLStradamus and cNLS mapper (available at http//nls-mapper.iab.keio.ac.jp/) programs. (B) Construction of expression plasmids containing various NLSs of GlMyb2. (C) Subcellular localization of full-length or truncated GlMYB2 in G. lamblia. GlMYB2 proteins were expressed from a constitutive gdh promoter in a HA tagged form. Giardia trophozoites attached to glass slides were reacted overnight with mouse anti HA (1:100) and then incubated with Alexa Fluor 488 conjugated anti mouse IgG (1:100). Differential interference contrast image was acquired to observe cell morphology. Scale bars=2 μm.
To examine and ascertain the putative NLS(s) required for the nuclear localization of GlMYB2, a series of plasmids, which expressed full-length GlMYB2 or truncated GlMYB2 losing of these putative NLS(s) in a hemagglutinin (HA)-tagged form, were constructed and transfected in Giardia trophozoites (Fig. 1B). A DNA fragment that contained the promoter for G. lamblia glutamate dehydrogenase (Glgdh) was constructed by PCR using primers, Pgdh-F and Pgdh-R (Supplement Table 1), and then cloned into pKS-3HA.neo [7] to obtain pPgdh-3HA. A full-length GlMYB2-encoding DNA fragment was amplified from Giardia genomic DNA using primers, Myb2-F and Myb2-R (Supplement Table 1), and cloned into plasmid pPgdh-3HA (Table 1). A PCR product, Myb21-498, was amplified using primers, Myb2-F and 498Myb2-R, and then used to express GlMYB2 without the third predicted NLS (#507-#530). The DNA fragments, Myb21-460 and Myb21-360, were prepared to obtain truncated GlMYB2 with the first and second NLSs and another truncated GlMYB2 with only the first NLS, respectively. Lastly, an additional expression plasmid was constructed to express truncated GlMYB2 without the NLSs (pPgdh-Myb21-329HA) in Giardia trophozoites.
The resulting plasmids were transfected into Giardia trophozoites by electroporation. G. lamblia trophozoites (WB; ATCC30957, American Type Culture Collection, Manassas, Virginia, USA) were grown in TYI-S-33 medium (2% casein digest, 1% yeast extract, 1% glucose, 0.2% NaCl, 0.2% L-cysteine, 0.02% ascorbic acid, 0.2% K2HPO4, 0.06% KH2PO4, 10% calf serum, and 0.5 mg/ml bovine bile, pH 7.1) at 37°C for 72 hr [8]. Twenty micrograms of plasmids were transfected into 1×107 Giardia trophozoites by electroporation under the following conditions: 350 V, 1,000 μF, and 700 Ω (Bio-Rad, Hercules, California, USA).
The resulting transgenic Giardia trophozoites were examined for localization of ectopically expressed GlMYB2-HA by immunofluorescence assays (IFA) using anti-HA antibodies (Fig. 1C). Giardia cells were attached on glass slides coated with L-lysine for 10 min, and then fixed with chilled methanol for 10 min, and phosphate buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4)/0.5% Triton X-100 for 10 min. After blocking in PBS/5% goat serum/3% bovine serum albumin for 1 hr, the cells were treated overnight with anti-HA mouse monoclonal antibodies (1:100; Sigma-Aldrich, St. Louis, Missouri, USA) at 4°C, and subsequently with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:100; Molecular Probes, Waltham, Massachusetts, USA). The slides were mounted with VECTASHIELD anti-fade mounting medium with DAPI (Vector Laboratories, Burlingame, California, USA), and examined with an Axiovert 200 fluorescent microscope (Carl Zeiss, Oberkochen, Germany).
Giardia cells that express full-length GlMYB2 (Pgdh-Myb21-530HA) demonstrated nuclear localization of GlMYB2 as expected [Fig. 1C upper panel center (Anti-HA)]. On the other hand, HA-tagged GlMYB21-498 (Pgdh-Myb21-498HA) was mainly found in the cytoplasm and rarely in the nuclei of the transgenic Giardia cells (Fig. 1C second panel center). The remaining Giardia cells expressing truncated GlMYB2 (Pgdh-Myb21-460HA, Pgdh-Myb21-360HA, Pgdh-Myb21-329HA) revealed the presence of these proteins in the cytoplasm (Fig. 1C). This result indicated that the third NLS (#507-#530) among the predicted NLSs of GlMYB2 (Fig. 1A) was required for nuclear localization of GlMYB2 in G. lamblia, and these sequences were named as NLSGlMYB2.
In the subsequent experiment, we verified the role of this NLS in nuclear localization using a chimeric protein obtained from a cytoplasmic protein, G. lamblia glyceraldehyde 3-phosphate dehydrogenase 1 (GlGAP1; GL50803_6687), and NLSGlMYB2 (Fig. 2). A GlGAP1-encoding PCR product was amplified from the Giardia genomic DNA using the primers, Gap1-F and Gap1-R (Supplementary Table S1), and cloned into pPgdh-3HA, resulting in pPgdh-GAP1 expressing GlGAP1 from the gdh promoter. A DNA fragment encoding mOrange was amplified from the plasmid, mOrange-pBAD (Addgene #54531, Watertown, Massachusetts, USA), and cloned into pPgdh-GAP1. The resulting pPgdh-GAP1-mOrange was used to express GlGAP1 fused with mOrange to facilitate microscopic observation without IFAs.
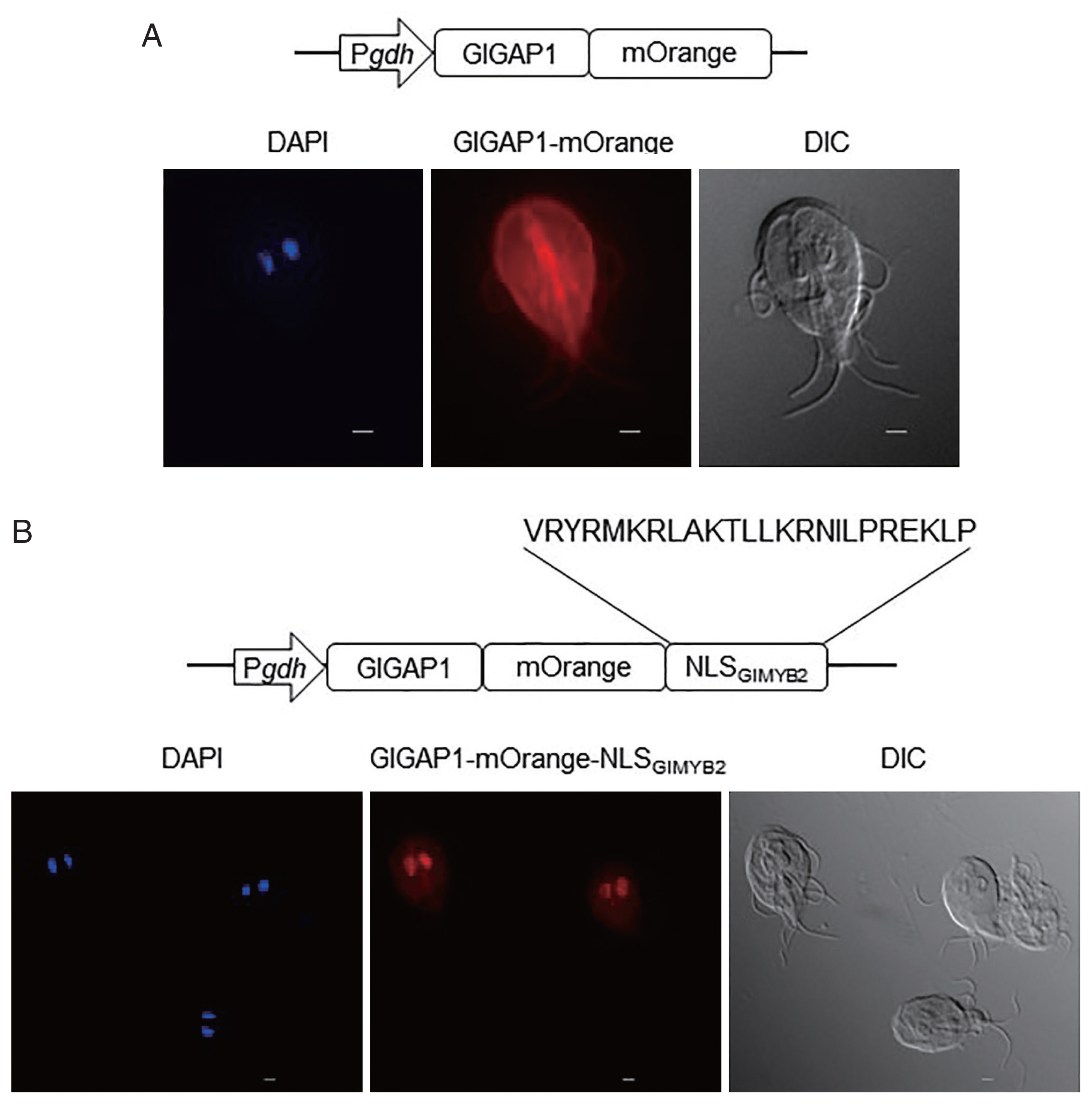
Role of the third nuclear localization signal (NLS) of G. lamblia MYB2 (GlMYB2) in the nuclear localization of cytoplasmic G. lamblia glyceraldehyde 3-phosphate dehydrogenase (GlGAP1). (A) A schematic diagram of the plasmid pPgdh-GAP1-mOrange. GlGAP1-mOrange was localized in cytoplasm and cytoskeletal structures (middle; orange color). DAPI represents the DNA in nuclei (left; blue color). (B) A schematic diagram of the plasmid pPgdh-GAP1-mOrange-NLSGlMYB2. This plasmid encodes NLS of GlMYB2 fused with mOrange. GlGAP1-mOrange-NLSGlMYB2 was localized in nuclei (middle; red color). The intracellular location of chimeric GlGAP1-mOrange was observed under a fluorescence microscope at 546 nm. Differential interference contrast image (DIC) was acquired to observe cell morphology. Scale bars=2 μm.
A PCR product encoding NLSGlMYB2 was cloned into the AflII and EcoRI sites of pPgdh-GAP1-mOrange, resulting in the plasmid expressing GlGAP1-mOrange-NLSMYB2. This plasmid was transfected into Giardia trophozoites as described above. The intracellular location of chimeric GlGAP1 protein was observed under the fluorescence microscope at 546 nm. When anti-GlGAP1 antibodies were used to observe the localization of GlGAP1 in Giardia trophozoite, GlGAP1 was expressed in cytoplasm except nuclei (Kim and Park, unpublished data). As expected, GlGAP1-mOrange was mainly observed in the cytoplasm and cytoskeletal structures (Fig. 2A center). If the NLSGlMYB2 is sufficient for translocation of the protein from the cytoplasm to the nuclei, GlGAP1-mOrange with NLSGlMYB2 would be expressed in nuclei. GlGAP1-mOrange-NLSGlMYB2 was found in the nuclei of Giardia trophozoites (Fig. 2B center). This result verified that the third NLS of GlMYB2 (#507-#530) was sufficient for nuclear localization of cytoplasmic protein GlGAP1 in Giardia trophozoites.
Nucleocytoplasmic transport is an essential process in eukaryotes and the machinery and mechanism involved in this process are conserved in organisms from yeasts to humans [9]. However, little is known about this process in protozoa. In Trypanosoma, classical NLSs found in other eukaryotes have been reported. Additionally, several nuclear proteins without this NLS indicate the presence of other complex mechanisms in Trypanosoma [10]. In G. lamblia, only the C-terminal NLS of the Giardia protein GL50803_2340 has been identified [6]. Our study provides experimental evidence on the NLS of the putative transcription factor, GlMYB2, which exerts its function during G. lamblia encystation.
Supplementary Information
Primers used in this study
ACKNOWLEDGMENT
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (NRF-2018R1D1A1A02085338 to S-J. Park and NRF-2020R1C1C1010581 to J. Kim)
Notes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.

