Clinical Usefulness of LabChip Real-Time PCR Using Lab-On-a-Chip Technology for Diagnosing Malaria
Article information
Abstract
As malaria remains a major health problem worldwide, various diagnostic tests have been developed, including microscopy-based and rapid diagnostic tests. LabChip real-time PCR (LRP) is a small and portable device used to diagnose malaria using lab-on-a-chip technology. This study aimed to evaluate the diagnostic performance of LRP for detecting malaria parasites. Two hundred thirteen patients and 150 healthy individuals were enrolled from May 2009 to October 2015. A diagnostic detectability of LRP for malaria parasites was compared to that of conventional RT-PCR. Sensitivity of LRP for Plasmodium vivax, P. falciparum, P. malariae, and P. ovale was 95.5%, 96.0%, 100%, and 100%, respectively. Specificity of LRP for P. vivax, P. falciparum, P. malariae, and P. ovale was 100%, 99.3%, 100%, and 100%, respectively. Cohen’s Kappa coefficients between LRP and CFX96 for detecting P. vivax, P. falciparum, P. malariae, and P. ovale were 0.96, 0.98, 1.00, and 1.00, respectively. Significant difference was not observed between the results of LRP and conventional RT-PCR and microscopic examination. A time required to amplify DNAs using LRP and conventional RT-PCR was 27 min and 86 min, respectively. LRP amplified DNAs 2 times more fast than conventional RT-PCR due to the faster heat transfer. Therefore, LRP could be employed as a useful tool for detecting malaria parasites in clinical laboratories.
Despite many initiatives by the World Health Organization and other agencies to eradicate malaria, it remains as a major health problem worldwide. About 40% of the world population lives in malaria-endemic areas, and 300–500 million cases are infected every year. In addition, approximately one million people die each year from malaria despite enormous investment in research and treatment projects to eradicate malaria [1,2]. In the Republic of Korea, the number of malaria infection cases decreased from 2,556 in 2001 to 559 in 2019, as the government and healthcare workers carried out various activities to treat and prevent malaria [3]. This decrease of malaria infections was largely due to early diagnosis and appropriate treatment. Since early diagnosis is important for the management of malaria patients, the development of fast and reliable diagnostic methods is very important. For these reasons, various programs have attempted to develop rapid and sensitive techniques for detecting malaria parasites.
Microscopic examination of Giemsa-stained blood smears is the gold standard method for detecting malaria parasites [4]. However, microscopic examination requires experts to detect malaria parasites and has several drawbacks such as subjective interpretation and the possibility of false negatives. In addition, accurate differential diagnosis can be difficult in the case of mixed malarial infections [5].
As an alternative to classical microscopic examination, many immunoassays have been developed. ICT Malaria Pf tests employed histidine-rich protein II antigen (HRP-2). OptiMAL tests employed Plasmodium-specific lactate dehydrogenase (pLDH). Various rapid diagnostic tests such as immunochromatographic tests, have a short turnaround time 15–20 min. Many clinicians use point-of-care test (POCT) diagnostic devices because these are easy to use and do not require special equipment. However, these POCTs showed lower sensitivity and specificity than the standard test methods [6]. Additionally, the prozone effect of HRP-2 based rapid diagnostic tests (RDTs) were reported to produce false-negative in samples from patients with hyperparasitemia [7]. In recent years, the P. falciparum deleted with PfHRP2 gene has been reported and responsible for increase of misdiagnosis by HRP2-based RDTs [8].
Recently, molecular diagnostic tests using PCR for DNA amplification have been used to diagnose malaria. Various molecular methods exhibit high sensitivity and specificity for malaria-parasite detection as they use species-specific primers for targeted DNA amplification [9]. However, molecular methods are not suitable for routine diagnosis of malaria-endemic areas due to their high cost, long turnaround times and the need for a well-equipped laboratory.
Microfluidic technologies have advanced dramatically over the years, particularly in the form of lab-on-a-chip (LOC) technology. These technologies have made it possible to miniaturize, integrate, and automate biochemical analyses [10]. Micro-electro-mechanical-system technologies were developed to enable faster and portable examination, and to enable miniature chip-type PCR. The advantages of LabChip real-time PCR include the following: shorter time for the reaction, requirement of a small reaction volume, consumption of fewer samples and reagents, and requirement of less power. The use of fewer samples and reagents for PCR could reduce the cost and allow rapid thermal cycling. Recently, LOC technology has been integrated with nucleic acid amplification methods to detect parasites responsible for causing infectious diseases [11]. LOC-based techniques are widely used to detect viruses such as Ebola, dengue, hepatitis B, human immunodeficiency viruses, and Mycobacterium tuberculosis [12–16]. These techniques have several advantages, including good portability, low sample consumption, short reaction time, compared to techniques requiring bulky benchtop equipment, indicating its potential application as a POCT.
LabChip real-time PCR (LRP; NanoBioSys, Seoul, Korea) is a newly developed real-time PCR system for malaria detection based on the analysis of the highly conserved 18S ribosomal RNA in patient samples. LRP is a compact portable system that uses LOC technology, and has advantages such as short PCR time, small equipment size, and fewer testing difficulties compared to conventional real-time PCR.
In this study, we compared diagnostic performance of LRP with that of microscopy-based examination and the CFX96 TouchTM Real-time PCR Detection System (CFX96; Bio-Rad, Hercules, California, USA), which is commonly used in clinical laboratories in the Republic of Korea. The purpose of this study was to evaluate clinical usefulness of LRP for detecting malaria parasites by analyzing the diagnostic performance of LRP.
From May 2009 to October 2015, a total of 363 individuals were enrolled, including 213 patients and 150 healthy individuals. This study was approved by the Institutional Review Board of Korea University Guro Hospital, Seoul, Republic of Korea (IRB No. 2017GR0785, 2020GR0451). A total of 163 blood samples were collected from patients positive for P. vivax, P. malariae, or P. ovale at the Korea University Guro Hospital. Fifty blood samples positive for P. falciparum were obtained from Myanmar in contractual agreement with Myanmar’s Health Service. Patients were confirmed for malarial infection based on the microscopy-based examination within 1 week of the symptom onset. The healthy individuals had not any history of malarial infection, nor traveled malaria-endemic areas in the past 3 years.
Whole blood samples were collected from 363 individuals in ethylenediaminetetraacetic acid (EDTA) tubes. All samples were stored at −80°C and tested by microscopic examination, LRP, and CFX96. Diagnostic performance of LRP and CFX96 for the malaria pararites were compared (Fig. 1).
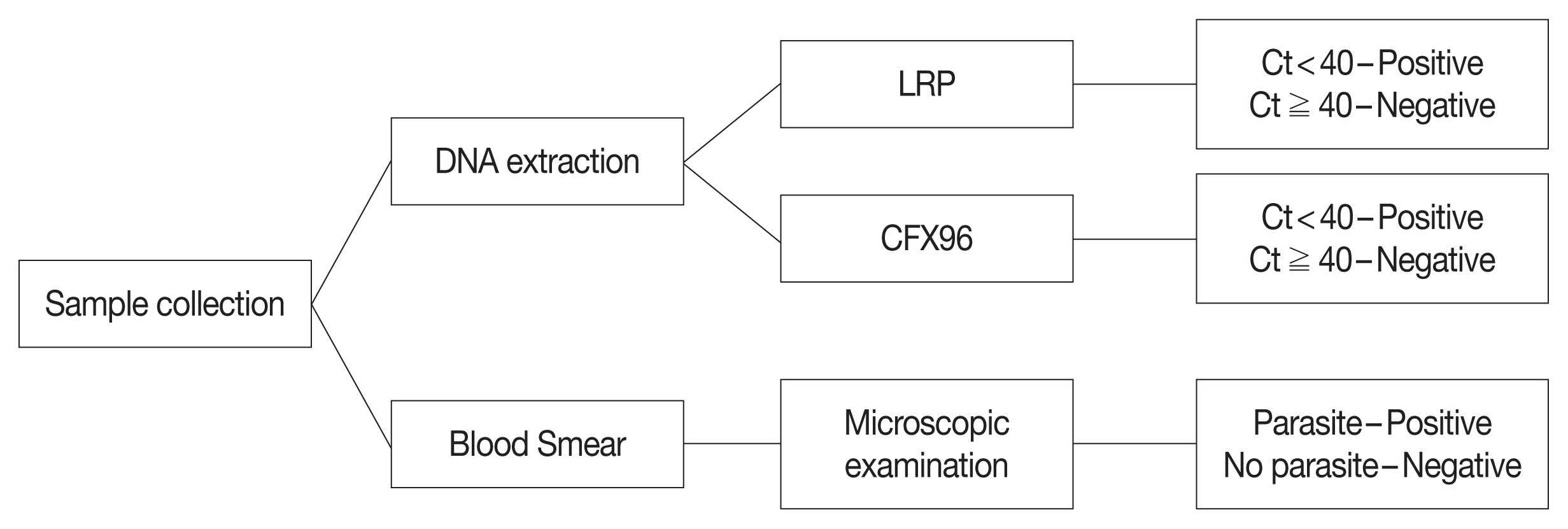
Flow chart of malarial parasites in blood samples. LRP, LabChip real-time PCR; CFX96, CFX96 touch real-time PCR; Ct, threadhold cycle.
Microscopic examination was used as a standard method for detecting the malaria parasites. For microscopy, thin and thick blood smears were prepared using standard protocols. Presence of the malaria parasites was determined by examining 200 high-power fields in blood smears. Malaria parasitemia level in the blood were calculated indirectly via microscopy as number of malaria parasites per 200 WBCs. WBC were counted using Coulter LH 780 Hematology Analyzer (Beckman Coulter Inc., Miami, Florida, USA).
Genomic DNA for LRP was isolated using the G2-16TU DNA extraction system (NanoBioSys). After mixing the genomic DNA with LRP malarial PCR master mix, 12 μl of the mixture was placed on the chip. The chip loaded with sample was inserted into the PCR machine, and LRP was conducted using specific primers against P. vivax, P. falciparum, P. malariae, and P. ovale. A test sample were determined positive when threshold cycle (Ct) value was less than 40, and negative if the Ct value was greater than or equal to 40.
Genomic DNA for CFX96 was extracted from peripheral blood using a DNA Extraction Kit (Qiagen Inc., Chatsworth, California, USA). Real-time PCR was performed according to the method previously reported [17], with minor modifications. Briefly, each tube contained a reaction volume of 20 μl comprising isolated DNA 2.5 μl, forward primer 0.1 μM, reverse primer 0.1 μM, and probe 0.1 μM. TaqMan amplification and detection were performed using CFX96. The following thermocycling conditions were used: 50°C for 2 min; 95°C for 10 min; 45 cycles of 95°C for 15 sec and 60°C for 1 min. Results were interpreted based on the Ct value when the normalized reporter dye emission exceeded the background noise. Test results were determined to be positive if the Ct value was less than 40 and negative if the Ct value was greater than or equal to 40.
Diagnostic performance of LRP was evaluated in terms of clinical sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and concordance rate. Sensitivity and specificity of LRP and CFX96 were assessed using microscopy, which is the standard method for detecting malaria parasites. Concordance rate between the LRP and CFX96 was calculated using inter-rater agreement statistics (kappa). A P-value of<0.05 was considered statistically significant. All statistical analyses were performed using SPSS for Windows (version 22.0; IBM Corporation, New York, New York, USA).
Characteristics of the patients included in this study: male to female patient ratio, 1.75 (231:132); mean age, 32±1.1 year, ranging 4 month to 71 year.
Of 363 samples, 157 were positive for P. vivax, 50 for P. falciparum, 4 for P. malariae, and 2 for P. ovale, by microscopic examination. Sensitivity of LRP for detecting P. vivax, P. falciparum, P. malariae, and P. ovale was 95.5%, 96.0%, 100%, and 100%, respectively. Sensitivity of CFX96 for P. vivax, P. falciparum, P. malariae, and P. ovale was 98.7%, 100%, 100%, and 100%, respectively. Specificities of LRP for P. vivax, P. falciparum, P. malariae, and P. ovale were 100%, 99.3%, 100%, and 100%, respectively. Specificity of CFX96 for P. vivax, P. falciparum, P. malariae, and P. ovale was 100%, 100%, 100%, and 100%, respectively (Table 1). PPV of LRP for detecting P. vivax, P. falciparum, P. malariae, and P. ovale was 100%, 100%, 100%, and 100%, respectively. NPV of LRP for detecting P. vivax, P. falciparum, P. malariae, and P. ovale was 96.71%, 99.4%, 100%, and 100%, respectively. Concordance rate between LRP and CFX96 for detecting P. vivax, P. falciparum, P. malariae, and P. ovale was 95.5%, 96.0%, 100%, and 100%, respectively. Cohen’s kappa coefficient between LRP and CFX96 for detecting P. vivax, P. falciparum, P. malariae, and P. ovale was 0.96, 0.98, 1.00, and 1.00, respectively (P<0.001,<0.001,<0.001, and <0.001, respectively). The McNemar’s test did not reveal significant difference between LRP and CFX96 for detecting P. vivax, P. falciparum, P. malariae, and P. ovale (P=0.125, 0.500, 1.000, and 1.000, respectively). The sensitivities of LRP and CFX96 based on P. vivax density are shown in Table 2. LRP presented a false negative result in a strongly positive sample at P. vivax density >5,000/μl, and 6 weakly positive samples at P. vivax density <500/μl. LRP and CFX96 Ct values were significantly correlated with parasite density, as assessed by microscopy (P<0.05). Correlation coefficients of LRP and CFX96 were 0.265 and 0.223, respectively.

Diagnostic performance of LabChip real-time PCR (LRP) and CFX96 Touch Real-time PCR (CFX96) compared to microscopic examination

Sensitivity of LabChip real-time PCR (LRP) and CFX96 Touch Real-time PCR assay according to density of Plasmodium vivax
Accurate and rapid malaria detection is clinically important for its treatment and prevention. Many molecular diagnostic methods, including nested PCR, loop-mediated isothermal amplification, and real-time PCR, have been developed to detect malaria parasites [18–20]. Compared to microscopic examination, overall sensitivity and specificity of commercial real-time PCR assays were 95.2–100% and 93.8–100%, respectively [21–23]. However, most real-time PCR assays are more complex and less intensive than conventional immunoassays, with longer testing times, making them unsuitable for routine use. Additionally, these tests can probably be misinterpreted as false positives if the samples are contaminated [24].
To compensate limits of these methods, LRP was developed as a portable chip-type real-time PCR analyzer weighing 5.5 kg. These chip-type real-time PCR assays have also shown high sensitivity and specificity for diagnosing influenza and tuberculosis [16,25]. LRP uses a chip-type real-time PCR technology to diagnose malaria with high sensitivity and specificity. The performance of real-time PCR coupled to lab-on-a-chip was evaluated and compared to that of standard real-time PCR tests to diagnose malaria. In this study, LRP exhibited a sensitivity of 95.5% and a specificity of 100%, similar to those of the conventional real-time PCR for detecting malaria parasites. Therefore, the diagnostic performance of LRP for detecting malaria parasites was comparable to that of CFX96.
LRP was designed to simplify the detection process and to enable rapid and accurate diagnosis by reducing the time required—from sampling to the confirmative diagnosis of malaria—within 1 hr 30 min. In this study, LRP took 27 min, while CFX96 took 86 min. Turn around time of LRP was more than 2 times fast than that of CFX96. Additionally, the results of LRP test were directly connected to the computer so that inspectors could easily interpret the results.
However, this study had several limitations. First, there were only 4 samples positive for P. malariae and 2 samples positive for P. ovale. Further studies should be conducted using more samples positive for P. malariae, P. ovale, and P. knowlesi to evaluate the clinical usefulness of LRP. It is also necessary to evaluate the efficiency of LRP against that of conventional real-time PCR assays. For detecting P. vivax, LRP showed false-negative results for 7 samples, and CFX96 showed a false-negative result for 2 samples. This could be attributed to low parasite density as chip-type real-time PCR is generally less efficient at extracting DNA than the real-time PCR assay commonly used in laboratories. This is because real-time PCR using LOC technology needs to be identified as having a slightly lower analytical sensitivity than conventional real-time PCR. Therefore, it is important to determine whether LRP is less efficient in extracting nucleic acids and whether there are other problems in detecting malaria parasites by amplifying nucleic acids. It will also be necessary to evaluate the detection limits of each method to ensure that LRP and CFX96 can diagnose malaria at an early stage.
In conclusion, this study demonstrated that LRP can rapidly detect malarial parasites with high sensitivity and specificity using LabChip technology. LRP was cost effective and amplifies nucleic acids 2 times faster than standard RT-PCR. It is expected that LRP will contribute to the diagnosis and control of malaria in endemic areas, especially in underdeveloped and developing countries.
ACKNOWLEDGMENTS
This study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health&Welfare, Republic of Korea (grant number: H15C1512, HR20C0021), a government-wide R&D fund project for infectious disease research (HG18C0012).
Notes
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interest.
